全文代码如下:
import mglearn
import joblib
import numpy as np
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
#线性回归
x,y = mglearn.datasets.make_wave(n_samples=60)
x_train,x_test,y_train,y_test = train_test_split(x,y,random_state=42)
lr = LinearRegression().fit(x_train,y_train)
print("lr.coef_:{}".format(lr.coef_))
print("lr.intercept_:{}".format(lr.intercept_))
print("training set score:{:.5f}".format(lr.score(x_train,y_train)))
print("test set score:{:.2f}".format(lr.score(x_test,y_test)))
#波士顿房价线性回归
x,y = mglearn.datasets.load_extended_boston()
x_train,x_test,y_train,y_test = train_test_split(x,y,random_state=0)
lr = LinearRegression().fit(x_train,y_train)
print("training set score:{:.2f}".format(lr.score(x_train,y_train)))
print("test set score:{:.2f}".format(lr.score(x_test,y_test)))
#岭回归
from sklearn.linear_model import Ridge
ridge = Ridge().fit(x_train,y_train)
print("train set score:{:.2f}".format(ridge.score(x_train,y_train)))
print("test set score:{:.2f}".format(ridge.score(x_test,y_test)))
ridge10 = Ridge(alpha=10).fit(x_train,y_train)
print("train set score:{:.2f}".format(ridge10.score(x_train,y_train)))
print("test set score:{:.2f}".format(ridge10.score(x_test,y_test)))
ridge01 = Ridge(alpha=0.1).fit(x_train,y_train)
print("train set score:{:.2f}".format(ridge01.score(x_train,y_train)))
print("test set score:{:.2f}".format(ridge01.score(x_test,y_test)))
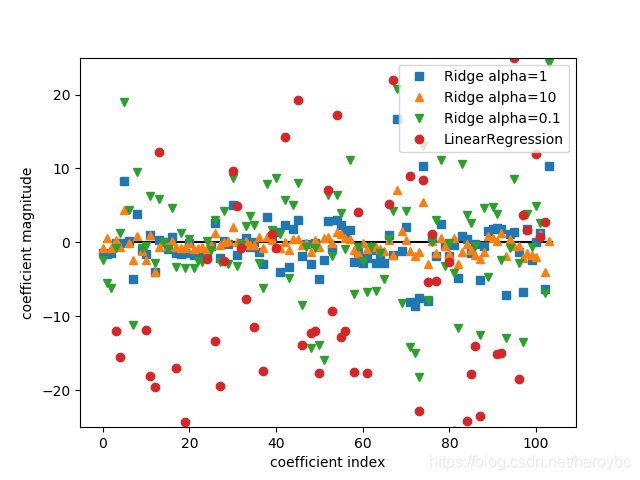
plt.plot(ridge.coef_,'s',label='Ridge alpha=1')
plt.plot(ridge10.coef_,'^',label='Ridge alpha=10')
plt.plot(ridge01.coef_,'v',label='Ridge alpha=0.1')
plt.plot(lr.coef_,'o',label='LinearRegression')
plt.xlabel('coefficient index')
plt.ylabel('coefficient magnitude')
plt.hlines(0,0,len(lr.coef_))
plt.ylim(-25,25)
plt.legend()
plt.show()
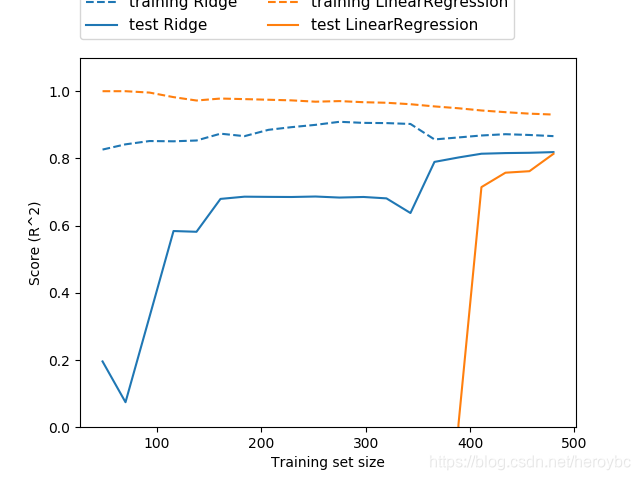
mglearn.plots.plot_ridge_n_samples()
plt.show()
#lasso
from sklearn.linear_model import Lasso
lasso = Lasso().fit(x_train,y_train)
print("train set score:{:.2f}".format(lasso.score(x_train,y_train)))
print("test set score:{:.2f}".format(lasso.score(x_test,y_test)))
print('number of feature used:{}'.format(np.sum(lasso.coef_ != 0)))
lasso001 = Lasso(alpha=0.01,max_iter=100000).fit(x_train,y_train)
print("train set score:{:.2f}".format(lasso001.score(x_train,y_train)))
print("test set score:{:.2f}".format(lasso001.score(x_test,y_test)))
print('number of feature used:{}'.format(np.sum(lasso001.coef_ != 0)))
lasso00001 = Lasso(alpha=0.0001,max_iter=100000).fit(x_train,y_train)
print("train set score:{:.2f}".format(lasso00001.score(x_train,y_train)))
print("test set score:{:.2f}".format(lasso00001.score(x_test,y_test)))
print('number of feature used:{}'.format(np.sum(lasso00001.coef_ != 0)))
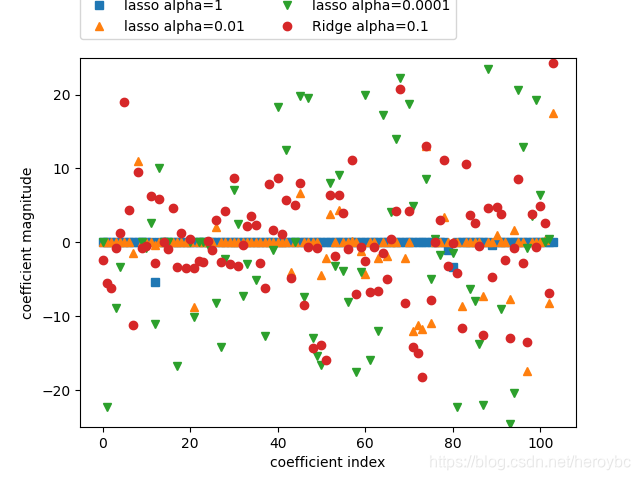
plt.plot(lasso.coef_,'s',label='lasso alpha=1')
plt.plot(lasso001.coef_,'^',label='lasso alpha=0.01')
plt.plot(lasso00001.coef_,'v',label='lasso alpha=0.0001')
plt.plot(ridge01.coef_,'o',label='Ridge alpha=0.1')
plt.xlabel('coefficient index')
plt.ylabel('coefficient magnitude')
plt.ylim(-25,25)
plt.legend(ncol=2,loc=(0,1.05))
plt.show()
#分类的线性模型
from sklearn.linear_model import LogisticRegression
from sklearn.svm import LinearSVC
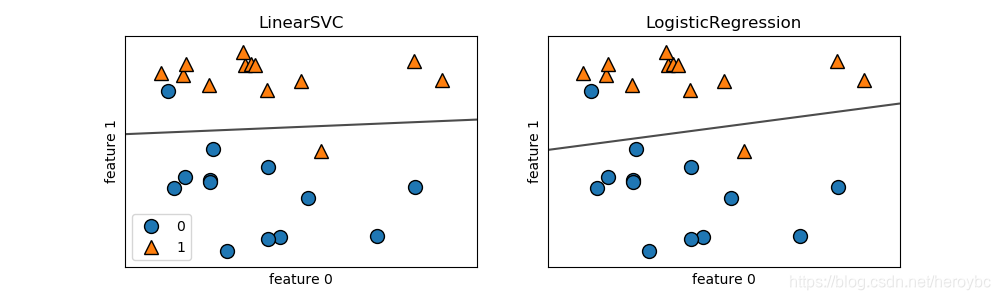
x,y = mglearn.datasets.make_forge()
fig,axes = plt.subplots(1,2,figsize=(10,3))
for model,ax in zip([LinearSVC(),LogisticRegression()],axes):
clf = model.fit(x,y)
mglearn.plots.plot_2d_separator(clf,x,fill=False,eps=0.5,ax=ax,alpha=.7)
mglearn.discrete_scatter(x[:,0],x[:,1],y,ax=ax)
ax.set_title('{}'.format(clf.__class__.__name__))
ax.set_xlabel('feature 0')
ax.set_ylabel('feature 1')
axes[0].legend()
plt.show()
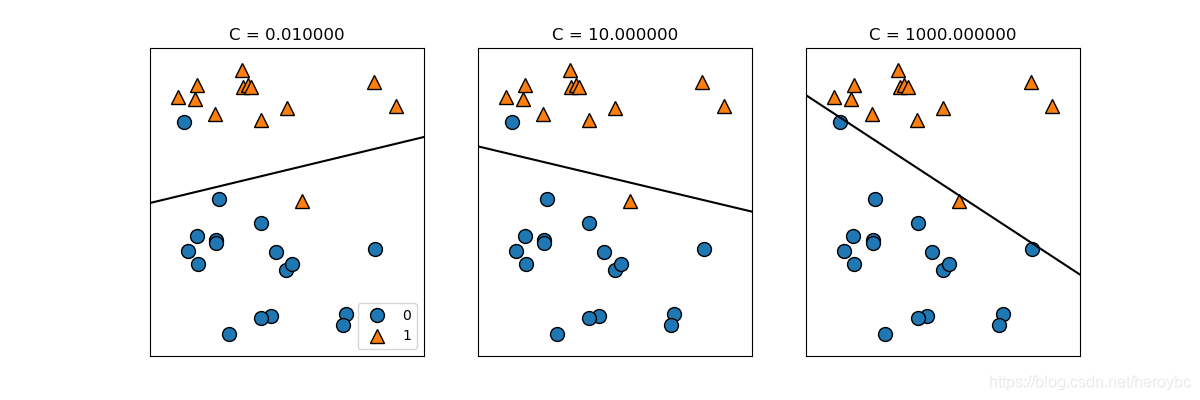
mglearn.plots.plot_linear_svc_regularization()
plt.show()
#在乳腺癌数据集分析逻辑回归(默认L2正则化)
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
x_train,x_test,y_train,y_test = train_test_split(cancer.data,cancer.target,stratify=cancer.target,random_state=42)
logreg = LogisticRegression().fit(x_train,y_train)
print("training set score:{:.3f}".format(logreg.score(x_train,y_train)))
print("test set score:{:.3f}".format(logreg.score(x_test,y_test)))
logreg100 = LogisticRegression(C=100).fit(x_train,y_train)
print("training set score:{:.3f}".format(logreg100.score(x_train,y_train)))
print("test set score:{:.3f}".format(logreg100.score(x_test,y_test)))
logreg001 = LogisticRegression(C=0.01).fit(x_train,y_train)
print("training set score:{:.3f}".format(logreg001.score(x_train,y_train)))
print("test set score:{:.3f}".format(logreg001.score(x_test,y_test)))
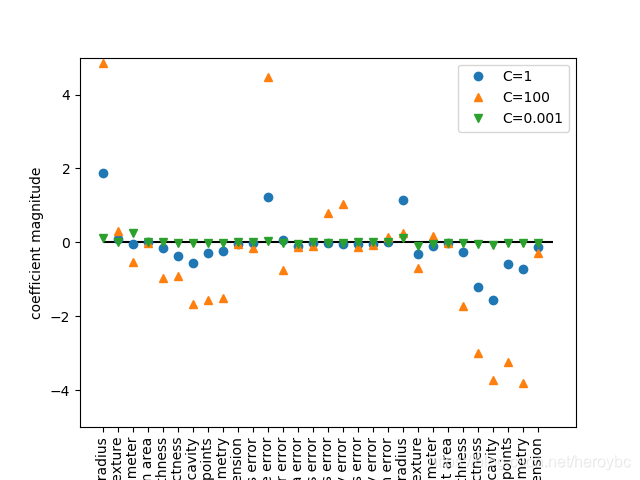
plt.plot(logreg.coef_.T,'o',label='C=1')
plt.plot(logreg100.coef_.T,'^',label='C=100')
plt.plot(logreg001.coef_.T,'v',label='C=0.001')
plt.xticks(range(cancer.data.shape[1]),cancer.feature_names,rotation=90)
plt.hlines(0,0,cancer.data.shape[1])
plt.ylim(-5,5)
plt.xlabel("coefficient index")
plt.ylabel("coefficient magnitude")
plt.legend()
plt.show()
#L1正则化
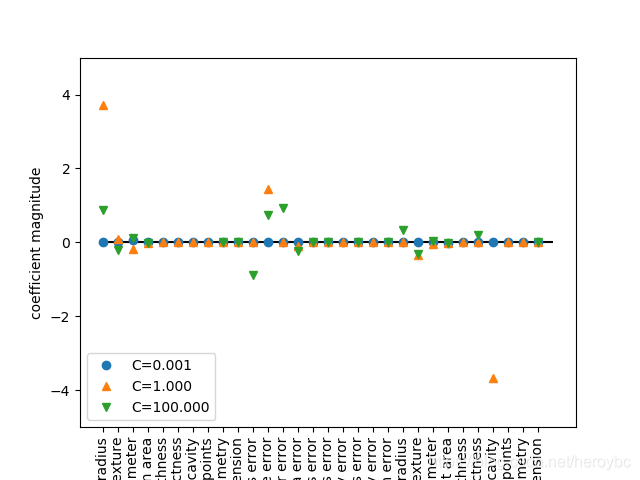
for C,marker in zip([0.001,1,100],['o','^','v']):
lr_l1 = LogisticRegression(C=C,penalty='l1').fit(x_train,y_train)
print('train accuracy of l1 logreg with C={:.3f}:{:.2f}'.format(C,lr_l1.score(x_train,y_train)))
print('test accuracy of l1 logreg with C={:.3f}:{:.2f}'.format(C,lr_l1.score(x_test,y_test)))
plt.plot(lr_l1.coef_.T,marker,label='C={:.3f}'.format(C))
plt.xticks(range(cancer.data.shape[1]),cancer.feature_names,rotation=90)
plt.hlines(0,0,cancer.data.shape[1])
plt.ylim(-5,5)
plt.xlabel("coefficient index")
plt.ylabel("coefficient magnitude")
plt.legend(loc=3)
plt.show()
#多分类的线性模型
from sklearn.datasets import make_blobs
from sklearn.svm import LinearSVC
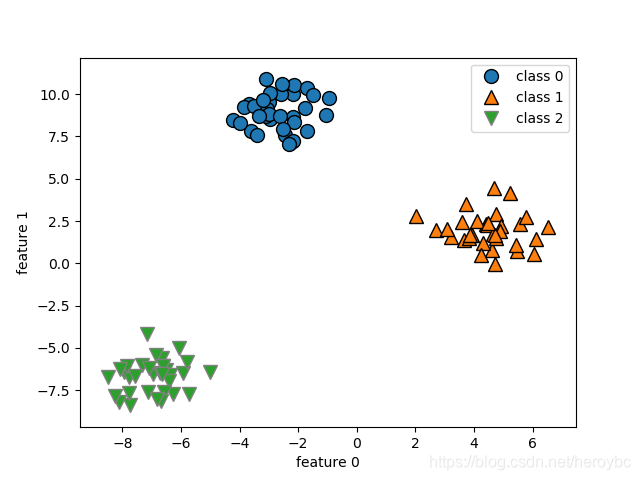
x,y = make_blobs(random_state=42)
mglearn.discrete_scatter(x[:,0],x[:,1],y)
plt.xlabel('feature 0')
plt.ylabel('feature 1')
plt.legend(['class 0','class 1','class 2'])
plt.show()
linear_svm = LinearSVC().fit(x,y)
print('coefficient shape:',linear_svm.coef_.shape)
print('intercept shape:',linear_svm.intercept_.shape)
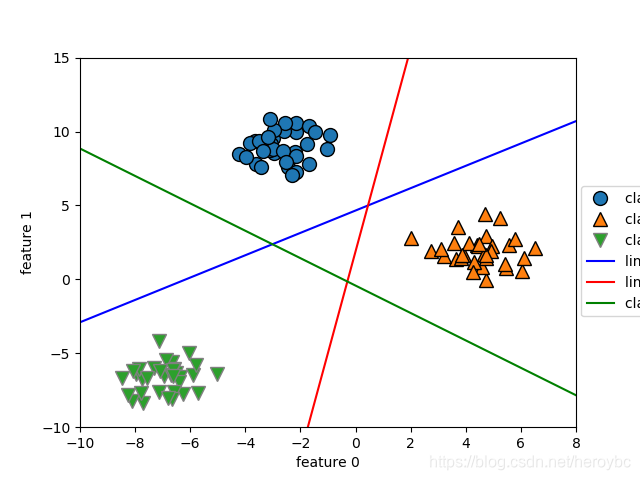
mglearn.discrete_scatter(x[:,0],x[:,1],y)
line = np.linspace(-15,15)
for coef,intercept,color in zip(linear_svm.coef_,linear_svm.intercept_,['b','r','g']):
plt.plot(line,-(line*coef[0]+intercept)/coef[1],c=color)
plt.ylim(-10,15)
plt.xlim(-10,8)
plt.xlabel('feature 0')
plt.ylabel('feature 1')
plt.legend(['class 0','class 1','class 2','line class 0','line class 1','class 2',],loc=(1.01,0.3))
plt.show()
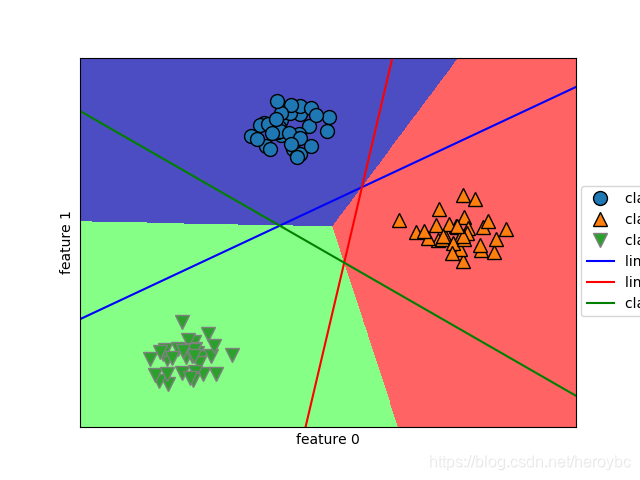
mglearn.plots.plot_2d_classification(linear_svm,x,fill=True,alpha=.7)
mglearn.discrete_scatter(x[:,0],x[:,1],y)
line = np.linspace(-15,15)
for coef,intercept,color in zip(linear_svm.coef_,linear_svm.intercept_,['b','r','g']):
plt.plot(line,-(line*coef[0]+intercept)/coef[1],c=color)
plt.legend(['class 0','class 1','class 2','line class 0','line class 1','class 2',],loc=(1.01,0.3))
plt.xlabel('feature 0')
plt.ylabel('feature 1')
plt.show()