用scikit-learn进行k-means聚类,默认使用欧式距离,为了用余弦距离作为度量,找了一个在生物信息学里比较常用的库:Biopython。Biopython为k-means聚类提供了各种距离函数,包括余弦距离、皮尔逊相似度量、欧式距离等。
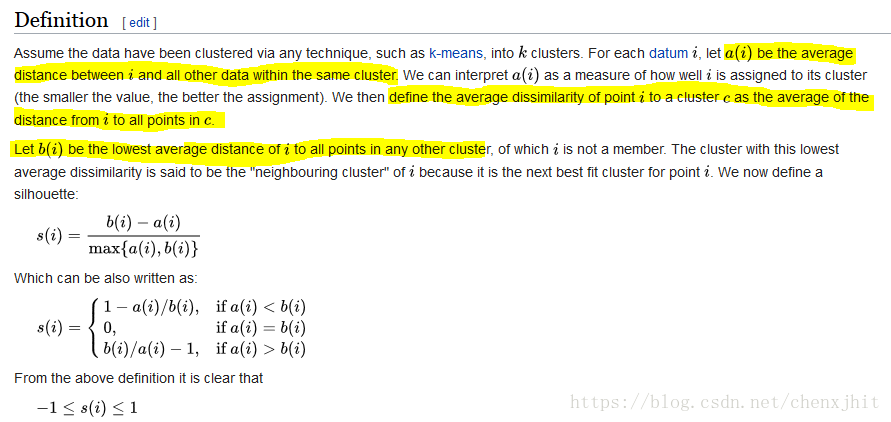
另外,为了确定一个合理的聚类系数,采用轮廓系数作为衡量标准:
轮廓系数取值为[-1, 1],其值越大越好。
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
from Bio.Cluster import kcluster
from Bio.Cluster import clustercentroids
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
data=np.load('/home/philochan/ResExp/genderkernel/1.npy')
coef = []
x=range(3,20)
for clusters in x:
clusterid, error, nfound = kcluster(data, clusters, dist='u',npass=100)
silhouette_avg = silhouette_score(data, clusterid, metric = 'cosine')
coef.append(silhouette_avg)
e =[i+3 for i,j in enumerate(coef) if j == max(coef)]
print e
print coef
plt.plot(x,coef)
plt.show()