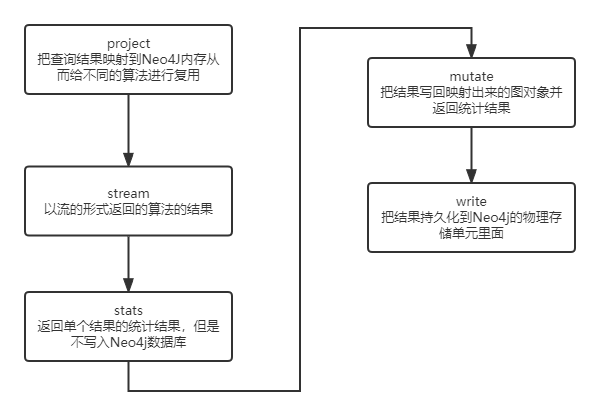
Neo4J 提供了GDS的库,里面包括了很多算法。GDS的英语全称是Graph Data Science(图数据科学库),其句法流程如下:
stream
Returns the result of the algorithm as a stream of records.
stats
Returns a single record of summary statistics, but does not write to the Neo4j database.
mutate
Writes the results of the algorithm to the projected graph and returns a single record of summary statistics.
write
Writes the results of the algorithm to the Neo4j database and returns a single record of summary statistics.
其提供的算法分成了下面8个种类。
-
Centrality 中心性算法
中心性算法用于确定网络中不同节点的重要性 -
Community detection 社区探测算法
社区检测算法用于评估节点组如何聚类或分区,以及它们加强或分裂的趋势 -
Similarity 相似性算法
相似度算法根据节点对的邻域或属性计算节点对的相似度。
可以使用几个相似性度量来计算相似性分数 -
Path finding 路径发现算法
路径查找算法查找两个或多个节点之间的路径或评估路径的可用性和质量 -
Node embeddings 节点嵌套算法
节点嵌入算法计算图中节点的低维向量表示。这些向量,也称为嵌入,可用于机器学习. -
Topological link prediction 拓扑链路预测算法
链路预测算法使用图的拓扑结构帮助确定一对节点的接近度。然后可以使用计算出的分数来预测它们之间的新关系 -
Auxiliary procedures 辅助流程算法
辅助程序是可以在您的工作流程中使用的额外工具。 -
Pregel API
Pregel 是一种以顶点为中心的计算模型,可通过用户定义的计算函数定义您自己的算法
默认情况下GDS库是没有安装的。如果需要安装,其安装步骤如下:
(1)需要GDS的仓库里面 然后复制到 N E O 4 J H O M E / p l u g i n s 目 录 下 。 ( 2 ) 在 NEO4J_HOME/plugins 目录下。 (2)在 NEO4JHOME/plugins目录下。(2)在NEO4J_HOME/conf/neo4j.conf文件里面修改下面的配置
dbms.security.procedures.unrestricted=gds.*
dbms.security.procedures.allowlist=gds.*
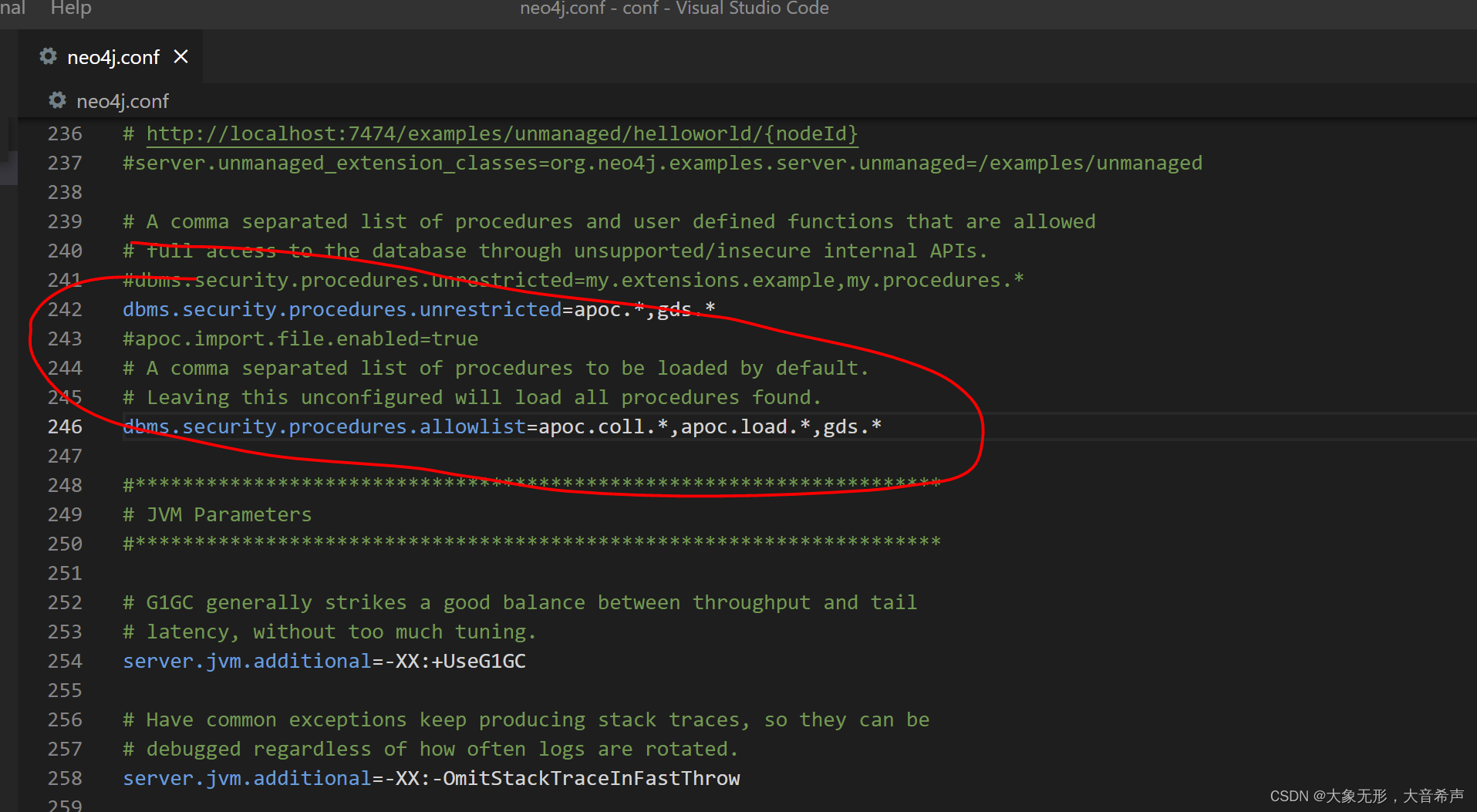
(3)重新启动Neo4J服务器
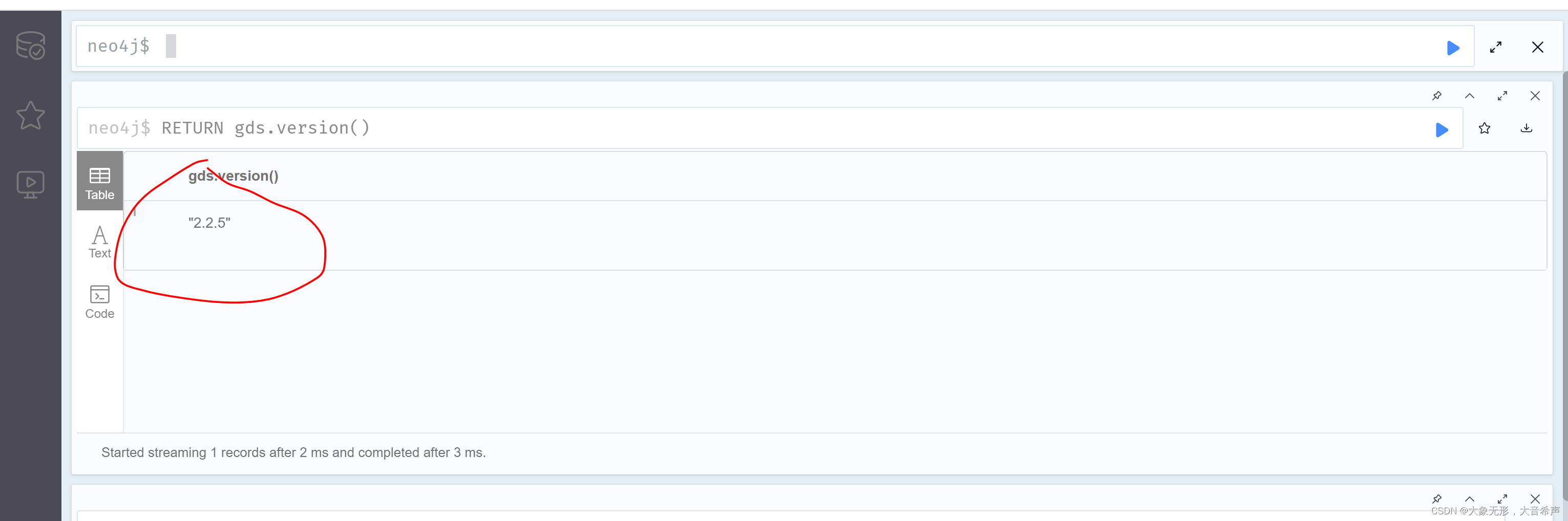
(4) 输入下面的命令进行验证
RETURN gds.version()
(5)查看其提供的函数
CALL gds.list()
[
{
"name": "gds.allShortestPaths.delta.mutate",
"description": "The Delta Stepping shortest path algorithm computes the shortest (weighted) path between one node and any other node in the graph. The computation is run multi-threaded",
"signature": "gds.allShortestPaths.delta.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.delta.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.allShortestPaths.delta.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.delta.stats",
"description": "The Delta Stepping shortest path algorithm computes the shortest (weighted) path between one node and any other node in the graph. The computation is run multi-threaded",
"signature": "gds.allShortestPaths.delta.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.delta.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.allShortestPaths.delta.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.delta.stream",
"description": "The Delta Stepping shortest path algorithm computes the shortest (weighted) path between one node and any other node in the graph. The computation is run multi-threaded",
"signature": "gds.allShortestPaths.delta.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (index :: INTEGER?, sourceNode :: INTEGER?, targetNode :: INTEGER?, totalCost :: FLOAT?, nodeIds :: LIST? OF INTEGER?, costs :: LIST? OF FLOAT?, path :: PATH?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.delta.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.allShortestPaths.delta.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.delta.write",
"description": "The Delta Stepping shortest path algorithm computes the shortest (weighted) path between one node and any other node in the graph. The computation is run multi-threaded",
"signature": "gds.allShortestPaths.delta.write(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.delta.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.allShortestPaths.delta.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.dijkstra.mutate",
"description": "The Dijkstra shortest path algorithm computes the shortest (weighted) path between one node and any other node in the graph.",
"signature": "gds.allShortestPaths.dijkstra.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.dijkstra.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.allShortestPaths.dijkstra.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.dijkstra.stream",
"description": "The Dijkstra shortest path algorithm computes the shortest (weighted) path between one node and any other node in the graph.",
"signature": "gds.allShortestPaths.dijkstra.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (index :: INTEGER?, sourceNode :: INTEGER?, targetNode :: INTEGER?, totalCost :: FLOAT?, nodeIds :: LIST? OF INTEGER?, costs :: LIST? OF FLOAT?, path :: PATH?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.dijkstra.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.allShortestPaths.dijkstra.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.dijkstra.write",
"description": "The Dijkstra shortest path algorithm computes the shortest (weighted) path between one node and any other node in the graph.",
"signature": "gds.allShortestPaths.dijkstra.write(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.allShortestPaths.dijkstra.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.allShortestPaths.dijkstra.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.allShortestPaths.stream",
"description": "The All Pairs Shortest Path (APSP) calculates the shortest (weighted) path between all pairs of nodes.",
"signature": "gds.alpha.allShortestPaths.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (sourceNodeId :: INTEGER?, targetNodeId :: INTEGER?, distance :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.backup",
"description": "The back-up procedure persists graphs and models to disk",
"signature": "gds.alpha.backup(configuration = {} :: MAP?) :: (graphName :: STRING?, modelName :: STRING?, backupTime :: DATETIME?, exportPath :: STRING?, exportMillis :: INTEGER?, status :: STRING?)",
"type": "procedure"
},
{
"name": "gds.alpha.closeness.harmonic.stream",
"description": "Harmonic centrality is a way of detecting nodes that are able to spread information very efficiently through a graph.",
"signature": "gds.alpha.closeness.harmonic.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, centrality :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.closeness.harmonic.write",
"description": "Harmonic centrality is a way of detecting nodes that are able to spread information very efficiently through a graph.",
"signature": "gds.alpha.closeness.harmonic.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodes :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, writeProperty :: STRING?, centralityDistribution :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.conductance.stream",
"description": "Evaluates a division of nodes into communities based on the proportion of relationships that cross community boundaries.",
"signature": "gds.alpha.conductance.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (community :: INTEGER?, conductance :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.config.defaults.list",
"description": "List defaults; global by default, but also optionally for a specific user and/ or key",
"signature": "gds.alpha.config.defaults.list(parameters = {} :: MAP?) :: (key :: STRING?, value :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.config.defaults.set",
"description": "Set a default; global by, default, but also optionally for a specific user",
"signature": "gds.alpha.config.defaults.set(key :: STRING?, value :: ANY?, username = d81eb72e-c499-4f78-90c7-0c76123606a2 :: STRING?) :: VOID",
"type": "procedure"
},
{
"name": "gds.alpha.config.limits.list",
"description": "List limits; global by default, but also optionally for a specific user and/ or key",
"signature": "gds.alpha.config.limits.list(parameters = {} :: MAP?) :: (key :: STRING?, value :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.config.limits.set",
"description": "Set a limit; global by, default, but also optionally for a specific user",
"signature": "gds.alpha.config.limits.set(key :: STRING?, value :: ANY?, username = d81eb72e-c499-4f78-90c7-0c76123606a2 :: STRING?) :: VOID",
"type": "procedure"
},
{
"name": "gds.alpha.create.cypherdb",
"description": "Creates a database from a GDS graph.",
"signature": "gds.alpha.create.cypherdb(dbName :: STRING?, graphName :: STRING?) :: (dbName :: STRING?, graphName :: STRING?, createMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.graph.graphProperty.drop",
"description": "Removes a graph property from a projected graph.",
"signature": "gds.alpha.graph.graphProperty.drop(graphName :: STRING?, graphProperty :: STRING?, configuration = {} :: MAP?) :: (graphName :: STRING?, graphProperty :: STRING?, propertiesRemoved :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.graph.graphProperty.stream",
"description": "Streams the given graph property.",
"signature": "gds.alpha.graph.graphProperty.stream(graphName :: STRING?, graphProperty :: STRING?, configuration = {} :: MAP?) :: (propertyValue :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.graph.sample.rwr",
"description": "Constructs a random subgraph based on random walks with restarts",
"signature": "gds.alpha.graph.sample.rwr(graphName :: STRING?, fromGraphName :: STRING?, configuration = {} :: MAP?) :: (fromGraphName :: STRING?, startNodeCount :: INTEGER?, graphName :: STRING?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, projectMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.hits.mutate",
"description": "Hyperlink-Induced Topic Search (HITS) is a link analysis algorithm that rates nodes",
"signature": "gds.alpha.hits.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.hits.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.hits.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.hits.stats",
"description": "Hyperlink-Induced Topic Search (HITS) is a link analysis algorithm that rates nodes",
"signature": "gds.alpha.hits.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.hits.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.hits.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.hits.stream",
"description": "Hyperlink-Induced Topic Search (HITS) is a link analysis algorithm that rates nodes",
"signature": "gds.alpha.hits.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, values :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.hits.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.hits.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.hits.write",
"description": "Hyperlink-Induced Topic Search (HITS) is a link analysis algorithm that rates nodes",
"signature": "gds.alpha.hits.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.hits.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.hits.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.influenceMaximization.greedy.stream",
"description": "The Greedy algorithm aims to find k nodes that maximize the expected spread of influence in the network.",
"signature": "gds.alpha.influenceMaximization.greedy.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, spread :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.kmeans.mutate",
"description": "The Kmeans algorithm clusters nodes into different communities based on Euclidean distance",
"signature": "gds.alpha.kmeans.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, communityDistribution :: MAP?, centroids :: LIST? OF LIST? OF FLOAT?, averageDistanceToCentroid :: FLOAT?, averageSilhouette :: FLOAT?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.kmeans.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.kmeans.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.kmeans.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.alpha.kmeans.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (communityDistribution :: MAP?, centroids :: LIST? OF LIST? OF FLOAT?, averageDistanceToCentroid :: FLOAT?, averageSilhouette :: FLOAT?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.kmeans.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.kmeans.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.kmeans.stream",
"description": "The Kmeans algorithm clusters nodes into different communities based on Euclidean distance",
"signature": "gds.alpha.kmeans.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, communityId :: INTEGER?, distanceFromCentroid :: FLOAT?, silhouette :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.kmeans.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.kmeans.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.kmeans.write",
"description": "The Kmeans algorithm clusters nodes into different communities based on Euclidean distance",
"signature": "gds.alpha.kmeans.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, communityDistribution :: MAP?, centroids :: LIST? OF LIST? OF FLOAT?, averageDistanceToCentroid :: FLOAT?, averageSilhouette :: FLOAT?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.kmeans.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.kmeans.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.knn.filtered.mutate",
"description": "The k-nearest neighbor graph algorithm constructs relationships between nodes if the distance between two nodes is among the k nearest distances compared to other nodes. KNN computes distances based on the similarity of node properties. Filtered KNN extends this functionality, allowing filtering on source nodes and target nodes, respectively.",
"signature": "gds.alpha.knn.filtered.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, nodePairsConsidered :: INTEGER?, didConverge :: BOOLEAN?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, relationshipsWritten :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.knn.filtered.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.alpha.knn.filtered.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, nodePairsConsidered :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, similarityPairs :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.knn.filtered.stream",
"description": "The k-nearest neighbor graph algorithm constructs relationships between nodes if the distance between two nodes is among the k nearest distances compared to other nodes. KNN computes distances based on the similarity of node properties. Filtered KNN extends this functionality, allowing filtering on source nodes and target nodes, respectively.",
"signature": "gds.alpha.knn.filtered.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (node1 :: INTEGER?, node2 :: INTEGER?, similarity :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.knn.filtered.write",
"description": "The k-nearest neighbor graph algorithm constructs relationships between nodes if the distance between two nodes is among the k nearest distances compared to other nodes. KNN computes distances based on the similarity of node properties. Filtered KNN extends this functionality, allowing filtering on source nodes and target nodes, respectively.",
"signature": "gds.alpha.knn.filtered.write(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, nodePairsConsidered :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, relationshipsWritten :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.leiden.mutate",
"description": "Leiden is a community detection algorithm, which guarantees that communities are well connected",
"signature": "gds.alpha.leiden.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranLevels :: INTEGER?, didConverge :: BOOLEAN?, nodeCount :: INTEGER?, communityCount :: INTEGER?, communityDistribution :: MAP?, modularity :: FLOAT?, modularities :: LIST? OF FLOAT?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.leiden.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.alpha.leiden.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranLevels :: INTEGER?, didConverge :: BOOLEAN?, nodeCount :: INTEGER?, communityCount :: INTEGER?, communityDistribution :: MAP?, modularity :: FLOAT?, modularities :: LIST? OF FLOAT?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.leiden.stream",
"description": "Leiden is a community detection algorithm, which guarantees that communities are well connected",
"signature": "gds.alpha.leiden.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, communityId :: INTEGER?, intermediateCommunityIds :: LIST? OF INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.leiden.write",
"description": "Leiden is a community detection algorithm, which guarantees that communities are well connected",
"signature": "gds.alpha.leiden.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranLevels :: INTEGER?, didConverge :: BOOLEAN?, nodeCount :: INTEGER?, communityCount :: INTEGER?, communityDistribution :: MAP?, modularity :: FLOAT?, modularities :: LIST? OF FLOAT?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.maxkcut.mutate",
"description": "Approximate Maximum k-cut maps each node into one of k disjoint communities trying to maximize the sum of weights of relationships between these communities.",
"signature": "gds.alpha.maxkcut.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, cutCost :: FLOAT?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.maxkcut.mutate.estimate",
"description": "Approximate Maximum k-cut maps each node into one of k disjoint communities trying to maximize the sum of weights of relationships between these communities.",
"signature": "gds.alpha.maxkcut.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.maxkcut.stream",
"description": "Approximate Maximum k-cut maps each node into one of k disjoint communities trying to maximize the sum of weights of relationships between these communities.",
"signature": "gds.alpha.maxkcut.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, communityId :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.maxkcut.stream.estimate",
"description": "Approximate Maximum k-cut maps each node into one of k disjoint communities trying to maximize the sum of weights of relationships between these communities.",
"signature": "gds.alpha.maxkcut.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.ml.splitRelationships.mutate",
"description": "Splits a graph into holdout and remaining relationship types and adds them to the graph.",
"signature": "gds.alpha.ml.splitRelationships.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, mutateMillis :: INTEGER?, relationshipsWritten :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.model.delete",
"description": "Deletes a stored model from disk.",
"signature": "gds.alpha.model.delete(modelName :: STRING?) :: (modelName :: STRING?, deleteMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.model.load",
"description": "Load a stored model into main memory.",
"signature": "gds.alpha.model.load(modelName :: STRING?) :: (modelName :: STRING?, loadMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.model.publish",
"description": "Make a trained model accessible by all users",
"signature": "gds.alpha.model.publish(modelName :: STRING?) :: (modelInfo :: MAP?, trainConfig :: MAP?, graphSchema :: MAP?, loaded :: BOOLEAN?, stored :: BOOLEAN?, creationTime :: DATETIME?, shared :: BOOLEAN?)",
"type": "procedure"
},
{
"name": "gds.alpha.model.store",
"description": "Store the selected model to disk.",
"signature": "gds.alpha.model.store(modelName :: STRING?, failIfUnsupportedType = true :: BOOLEAN?) :: (modelName :: STRING?, storeMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.modularity.stats",
"description": "TODO: Add modularity description",
"signature": "gds.alpha.modularity.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeCount :: INTEGER?, relationshipCount :: INTEGER?, communityCount :: INTEGER?, modularity :: FLOAT?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.modularity.stream",
"description": "TODO: Add modularity description",
"signature": "gds.alpha.modularity.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (communityId :: INTEGER?, modularity :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.nodeSimilarity.filtered.mutate",
"description": "The Filtered Node Similarity algorithm compares a set of nodes based on the nodes they are connected to. Two nodes are considered similar if they share many of the same neighbors. The algorithm computes pair-wise similarities based on Jaccard or Overlap metrics. The filtered variant supports limiting which nodes to compare via source and target node filters.",
"signature": "gds.alpha.nodeSimilarity.filtered.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, relationshipsWritten :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.nodeSimilarity.filtered.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.nodeSimilarity.filtered.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.nodeSimilarity.filtered.stats",
"description": "The Filtered Node Similarity algorithm compares a set of nodes based on the nodes they are connected to. Two nodes are considered similar if they share many of the same neighbors. The algorithm computes pair-wise similarities based on Jaccard or Overlap metrics. The filtered variant supports limiting which nodes to compare via source and target node filters.",
"signature": "gds.alpha.nodeSimilarity.filtered.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, similarityPairs :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.nodeSimilarity.filtered.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.nodeSimilarity.filtered.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.nodeSimilarity.filtered.stream",
"description": "The Filtered Node Similarity algorithm compares a set of nodes based on the nodes they are connected to. Two nodes are considered similar if they share many of the same neighbors. The algorithm computes pair-wise similarities based on Jaccard or Overlap metrics. The filtered variant supports limiting which nodes to compare via source and target node filters.",
"signature": "gds.alpha.nodeSimilarity.filtered.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (node1 :: INTEGER?, node2 :: INTEGER?, similarity :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.nodeSimilarity.filtered.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.nodeSimilarity.filtered.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.nodeSimilarity.filtered.write",
"description": "The Filtered Node Similarity algorithm compares a set of nodes based on the nodes they are connected to. Two nodes are considered similar if they share many of the same neighbors. The algorithm computes pair-wise similarities based on Jaccard or Overlap metrics. The filtered variant supports limiting which nodes to compare via source and target node filters.",
"signature": "gds.alpha.nodeSimilarity.filtered.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, relationshipsWritten :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.nodeSimilarity.filtered.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.nodeSimilarity.filtered.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.linkPrediction.addMLP",
"description": "Add a multilayer perceptron configuration to the parameter space of the link prediction train pipeline.",
"signature": "gds.alpha.pipeline.linkPrediction.addMLP(pipelineName :: STRING?, config = {} :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureSteps :: LIST? OF MAP?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.linkPrediction.addRandomForest",
"description": "Add a random forest configuration to the parameter space of the link prediction train pipeline.",
"signature": "gds.alpha.pipeline.linkPrediction.addRandomForest(pipelineName :: STRING?, config :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureSteps :: LIST? OF MAP?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.linkPrediction.configureAutoTuning",
"description": "Configures the auto-tuning of the link prediction pipeline.",
"signature": "gds.alpha.pipeline.linkPrediction.configureAutoTuning(pipelineName :: STRING?, configuration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureSteps :: LIST? OF MAP?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeClassification.addMLP",
"description": "Add a multilayer perceptron configuration to the parameter space of the node classification train pipeline.",
"signature": "gds.alpha.pipeline.nodeClassification.addMLP(pipelineName :: STRING?, config = {} :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeClassification.addRandomForest",
"description": "Add a random forest configuration to the parameter space of the node classification train pipeline.",
"signature": "gds.alpha.pipeline.nodeClassification.addRandomForest(pipelineName :: STRING?, config :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeClassification.configureAutoTuning",
"description": "Configures the auto-tuning of the node classification pipeline.",
"signature": "gds.alpha.pipeline.nodeClassification.configureAutoTuning(pipelineName :: STRING?, configuration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.addLinearRegression",
"description": "Add a linear regression model candidate to a node regression pipeline.",
"signature": "gds.alpha.pipeline.nodeRegression.addLinearRegression(pipelineName :: STRING?, configuration = {} :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.addNodeProperty",
"description": "Add a node property step to an existing node regression training pipeline.",
"signature": "gds.alpha.pipeline.nodeRegression.addNodeProperty(pipelineName :: STRING?, procedureName :: STRING?, procedureConfiguration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.addRandomForest",
"description": "Add a random forest model candidate to a node regression pipeline.",
"signature": "gds.alpha.pipeline.nodeRegression.addRandomForest(pipelineName :: STRING?, configuration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.configureAutoTuning",
"description": "Configures the auto-tuning of a node regression pipeline.",
"signature": "gds.alpha.pipeline.nodeRegression.configureAutoTuning(pipelineName :: STRING?, configuration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.configureSplit",
"description": "Configures the graph splitting of a node regression pipeline.",
"signature": "gds.alpha.pipeline.nodeRegression.configureSplit(pipelineName :: STRING?, configuration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.create",
"description": "Creates a node regression training pipeline in the pipeline catalog.",
"signature": "gds.alpha.pipeline.nodeRegression.create(pipelineName :: STRING?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.predict.mutate",
"description": "Predicts target node property using a previously trained `NodeRegression` model",
"signature": "gds.alpha.pipeline.nodeRegression.predict.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.predict.stream",
"description": "Predicts target node property using a previously trained `NodeRegression` model",
"signature": "gds.alpha.pipeline.nodeRegression.predict.stream(graphName :: STRING?, configuration :: MAP?) :: (nodeId :: INTEGER?, predictedValue :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.selectFeatures",
"description": "Add one or several features to an existing node regression training pipeline.",
"signature": "gds.alpha.pipeline.nodeRegression.selectFeatures(pipelineName :: STRING?, featureProperties :: ANY?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.alpha.pipeline.nodeRegression.train",
"description": "Trains a node classification model based on a pipeline",
"signature": "gds.alpha.pipeline.nodeRegression.train(graphName :: STRING?, configuration = {} :: MAP?) :: (modelSelectionStats :: MAP?, trainMillis :: INTEGER?, modelInfo :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.restore",
"description": "The restore procedure reads graphs and models from disk.",
"signature": "gds.alpha.restore(configuration = {} :: MAP?) :: (restoredGraph :: STRING?, restoredModel :: STRING?, status :: STRING?, restoreMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.scaleProperties.mutate",
"description": "Scale node properties",
"signature": "gds.alpha.scaleProperties.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.scaleProperties.stream",
"description": "Scale node properties",
"signature": "gds.alpha.scaleProperties.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, scaledProperty :: LIST? OF FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.scc.stream",
"description": "The SCC algorithm finds sets of connected nodes in an directed graph, where all nodes in the same set form a connected component.",
"signature": "gds.alpha.scc.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, componentId :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.scc.write",
"description": "The SCC algorithm finds sets of connected nodes in an directed graph, where all nodes in the same set form a connected component.",
"signature": "gds.alpha.scc.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodes :: INTEGER?, communityCount :: INTEGER?, setCount :: INTEGER?, minSetSize :: INTEGER?, maxSetSize :: INTEGER?, p1 :: INTEGER?, p5 :: INTEGER?, p10 :: INTEGER?, p25 :: INTEGER?, p50 :: INTEGER?, p75 :: INTEGER?, p90 :: INTEGER?, p95 :: INTEGER?, p99 :: INTEGER?, p100 :: INTEGER?, writeProperty :: STRING?)",
"type": "procedure"
},
{
"name": "gds.alpha.sllpa.mutate",
"description": "The Speaker Listener Label Propagation algorithm is a fast algorithm for finding overlapping communities in a graph.",
"signature": "gds.alpha.sllpa.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.sllpa.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.sllpa.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.sllpa.stats",
"description": "The Speaker Listener Label Propagation algorithm is a fast algorithm for finding overlapping communities in a graph.",
"signature": "gds.alpha.sllpa.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.sllpa.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.sllpa.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.sllpa.stream",
"description": "The Speaker Listener Label Propagation algorithm is a fast algorithm for finding overlapping communities in a graph.",
"signature": "gds.alpha.sllpa.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, values :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.sllpa.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.sllpa.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.sllpa.write",
"description": "The Speaker Listener Label Propagation algorithm is a fast algorithm for finding overlapping communities in a graph.",
"signature": "gds.alpha.sllpa.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.sllpa.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.alpha.sllpa.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.spanningTree.kmax.write",
"description": "The maximum weight spanning tree (MST) starts from a given node, and finds all its reachable nodes and the set of relationships that connect the nodes together with the maximum possible weight.",
"signature": "gds.alpha.spanningTree.kmax.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, effectiveNodeCount :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.spanningTree.kmin.write",
"description": "The minimum weight spanning tree (MST) starts from a given node, and finds all its reachable nodes and the set of relationships that connect the nodes together with the minimum possible weight.",
"signature": "gds.alpha.spanningTree.kmin.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, effectiveNodeCount :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.spanningTree.maximum.write",
"description": "Maximum weight spanning tree visits all nodes that are in the same connected component as the starting node, and returns a spanning tree of all nodes in the component where the total weight of the relationships is maximized.",
"signature": "gds.alpha.spanningTree.maximum.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, effectiveNodeCount :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.spanningTree.minimum.write",
"description": "Minimum weight spanning tree visits all nodes that are in the same connected component as the starting node, and returns a spanning tree of all nodes in the component where the total weight of the relationships is minimized.",
"signature": "gds.alpha.spanningTree.minimum.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, effectiveNodeCount :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.spanningTree.write",
"description": "Minimum weight spanning tree visits all nodes that are in the same connected component as the starting node, and returns a spanning tree of all nodes in the component where the total weight of the relationships is minimized.",
"signature": "gds.alpha.spanningTree.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, effectiveNodeCount :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.systemMonitor",
"description": "Get an overview of the system's workload and available resources",
"signature": "gds.alpha.systemMonitor() :: (freeHeap :: INTEGER?, totalHeap :: INTEGER?, maxHeap :: INTEGER?, jvmAvailableCpuCores :: INTEGER?, availableCpuCoresNotRequested :: INTEGER?, jvmHeapStatus :: MAP?, ongoingGdsProcedures :: LIST? OF MAP?)",
"type": "procedure"
},
{
"name": "gds.alpha.triangles",
"description": "Triangles streams the nodeIds of each triangle in the graph.",
"signature": "gds.alpha.triangles(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeA :: INTEGER?, nodeB :: INTEGER?, nodeC :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.alpha.userLog",
"description": "Log warnings and hints for currently running tasks.",
"signature": "gds.alpha.userLog(jobId = :: STRING?) :: (taskName :: STRING?, message :: STRING?, timeStarted :: LOCALTIME?)",
"type": "procedure"
},
{
"name": "gds.articleRank.mutate",
"description": "Article Rank is a variant of the Page Rank algorithm, which measures the transitive influence or connectivity of nodes.",
"signature": "gds.articleRank.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.articleRank.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.articleRank.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.articleRank.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.articleRank.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.articleRank.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.articleRank.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.articleRank.stream",
"description": "Article Rank is a variant of the Page Rank algorithm, which measures the transitive influence or connectivity of nodes.",
"signature": "gds.articleRank.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, score :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.articleRank.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.articleRank.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.articleRank.write",
"description": "Article Rank is a variant of the Page Rank algorithm, which measures the transitive influence or connectivity of nodes.",
"signature": "gds.articleRank.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.articleRank.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.articleRank.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.closeness.mutate",
"description": "Closeness centrality is a way of detecting nodes that are able to spread information very efficiently through a graph.",
"signature": "gds.beta.closeness.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, mutateProperty :: STRING?, centralityDistribution :: MAP?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.closeness.stats",
"description": "Closeness centrality is a way of detecting nodes that are able to spread information very efficiently through a graph.",
"signature": "gds.beta.closeness.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.closeness.stream",
"description": "Closeness centrality is a way of detecting nodes that are able to spread information very efficiently through a graph.",
"signature": "gds.beta.closeness.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, score :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.closeness.write",
"description": "Closeness centrality is a way of detecting nodes that are able to spread information very efficiently through a graph.",
"signature": "gds.beta.closeness.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, writeProperty :: STRING?, centralityDistribution :: MAP?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.collapsePath.mutate",
"description": "Collapse Path algorithm is a traversal algorithm capable of creating relationships between the start and end nodes of a traversal",
"signature": "gds.beta.collapsePath.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, mutateMillis :: INTEGER?, relationshipsWritten :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.graph.export.csv",
"description": "Exports a named graph to CSV files.",
"signature": "gds.beta.graph.export.csv(graphName :: STRING?, configuration = {} :: MAP?) :: (exportName :: STRING?, graphName :: STRING?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, relationshipTypeCount :: INTEGER?, nodePropertyCount :: INTEGER?, relationshipPropertyCount :: INTEGER?, writeMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.beta.graph.export.csv.estimate",
"description": "Estimate the required disk space for exporting a named graph to CSV files.",
"signature": "gds.beta.graph.export.csv.estimate(graphName :: STRING?, configuration = {} :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.graph.generate",
"description": "Computes a random graph, which will be stored in the graph catalog.",
"signature": "gds.beta.graph.generate(graphName :: STRING?, nodeCount :: INTEGER?, averageDegree :: INTEGER?, configuration = {} :: MAP?) :: (name :: STRING?, nodes :: INTEGER?, relationships :: INTEGER?, generateMillis :: INTEGER?, relationshipSeed :: INTEGER?, averageDegree :: FLOAT?, relationshipDistribution :: ANY?, relationshipProperty :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.graph.project.subgraph",
"description": "Creates a named graph in the catalog for use by algorithms.",
"signature": "gds.beta.graph.project.subgraph(graphName :: STRING?, fromGraphName :: STRING?, nodeFilter :: STRING?, relationshipFilter :: STRING?, configuration = {} :: MAP?) :: (fromGraphName :: STRING?, nodeFilter :: STRING?, relationshipFilter :: STRING?, graphName :: STRING?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, projectMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.beta.graph.relationships.stream",
"description": "Streams the given relationship source/target pairs",
"signature": "gds.beta.graph.relationships.stream(graphName :: STRING?, relationshipTypes = [*] :: LIST? OF STRING?, configuration = {} :: MAP?) :: (sourceNodeId :: INTEGER?, targetNodeId :: INTEGER?, relationshipType :: STRING?)",
"type": "procedure"
},
{
"name": "gds.beta.graphSage.mutate",
"description": "The GraphSage algorithm inductively computes embeddings for nodes based on a their features and neighborhoods.",
"signature": "gds.beta.graphSage.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, mutateMillis :: INTEGER?, nodeCount :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.graphSage.mutate.estimate",
"description": "The GraphSage algorithm inductively computes embeddings for nodes based on a their features and neighborhoods.",
"signature": "gds.beta.graphSage.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.graphSage.stream",
"description": "The GraphSage algorithm inductively computes embeddings for nodes based on a their features and neighborhoods.",
"signature": "gds.beta.graphSage.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, embedding :: LIST? OF FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.graphSage.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.graphSage.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.graphSage.train",
"description": "The GraphSage algorithm inductively computes embeddings for nodes based on a their features and neighborhoods.",
"signature": "gds.beta.graphSage.train(graphName :: STRING?, configuration = {} :: MAP?) :: (modelInfo :: MAP?, configuration :: MAP?, trainMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.beta.graphSage.train.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.graphSage.train.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.graphSage.write",
"description": "The GraphSage algorithm inductively computes embeddings for nodes based on a their features and neighborhoods.",
"signature": "gds.beta.graphSage.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeCount :: INTEGER?, nodePropertiesWritten :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.graphSage.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.graphSage.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.influenceMaximization.celf.mutate",
"description": "The Cost Effective Lazy Forward (CELF) algorithm aims to find k nodes that maximize the expected spread of influence in the network.",
"signature": "gds.beta.influenceMaximization.celf.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, computeMillis :: INTEGER?, totalSpread :: FLOAT?, nodeCount :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.influenceMaximization.celf.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.influenceMaximization.celf.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.influenceMaximization.celf.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.beta.influenceMaximization.celf.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (computeMillis :: INTEGER?, totalSpread :: FLOAT?, nodeCount :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.influenceMaximization.celf.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.influenceMaximization.celf.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.influenceMaximization.celf.stream",
"description": "The Cost Effective Lazy Forward (CELF) algorithm aims to find k nodes that maximize the expected spread of influence in the network.",
"signature": "gds.beta.influenceMaximization.celf.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, spread :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.influenceMaximization.celf.stream.estimate",
"description": "The Cost Effective Lazy Forward (CELF) algorithm aims to find k nodes that maximize the expected spread of influence in the network.",
"signature": "gds.beta.influenceMaximization.celf.stream.estimate(graphName :: ANY?, configuration = {} :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.influenceMaximization.celf.write",
"description": "The Cost Effective Lazy Forward (CELF) algorithm aims to find k nodes that maximize the expected spread of influence in the network.",
"signature": "gds.beta.influenceMaximization.celf.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, computeMillis :: INTEGER?, totalSpread :: FLOAT?, nodeCount :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.influenceMaximization.celf.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.influenceMaximization.celf.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.k1coloring.mutate",
"description": "The K-1 Coloring algorithm assigns a color to every node in the graph.",
"signature": "gds.beta.k1coloring.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, mutateMillis :: INTEGER?, nodeCount :: INTEGER?, colorCount :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.k1coloring.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.k1coloring.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.k1coloring.stats",
"description": "The K-1 Coloring algorithm assigns a color to every node in the graph.",
"signature": "gds.beta.k1coloring.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, nodeCount :: INTEGER?, colorCount :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.k1coloring.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.k1coloring.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.k1coloring.stream",
"description": "The K-1 Coloring algorithm assigns a color to every node in the graph.",
"signature": "gds.beta.k1coloring.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, color :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.beta.k1coloring.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.k1coloring.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.k1coloring.write",
"description": "The K-1 Coloring algorithm assigns a color to every node in the graph.",
"signature": "gds.beta.k1coloring.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, nodeCount :: INTEGER?, colorCount :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.k1coloring.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.k1coloring.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.listProgress",
"description": "List progress events for currently running tasks.",
"signature": "gds.beta.listProgress(jobId = :: STRING?) :: (jobId :: STRING?, taskName :: STRING?, progress :: STRING?, progressBar :: STRING?, status :: STRING?, timeStarted :: LOCALTIME?, elapsedTime :: STRING?)",
"type": "procedure"
},
{
"name": "gds.beta.model.drop",
"description": "Drops a loaded model and frees up the resources it occupies.",
"signature": "gds.beta.model.drop(modelName :: STRING?, failIfMissing = true :: BOOLEAN?) :: (modelInfo :: MAP?, trainConfig :: MAP?, graphSchema :: MAP?, loaded :: BOOLEAN?, stored :: BOOLEAN?, creationTime :: DATETIME?, shared :: BOOLEAN?)",
"type": "procedure"
},
{
"name": "gds.beta.model.exists",
"description": "Checks if a given model exists in the model catalog.",
"signature": "gds.beta.model.exists(modelName :: STRING?) :: (modelName :: STRING?, modelType :: STRING?, exists :: BOOLEAN?)",
"type": "procedure"
},
{
"name": "gds.beta.model.list",
"description": "Lists all models contained in the model catalog.",
"signature": "gds.beta.model.list(modelName = __NO_VALUE :: STRING?) :: (modelInfo :: MAP?, trainConfig :: MAP?, graphSchema :: MAP?, loaded :: BOOLEAN?, stored :: BOOLEAN?, creationTime :: DATETIME?, shared :: BOOLEAN?)",
"type": "procedure"
},
{
"name": "gds.beta.modularityOptimization.mutate",
"description": "The Modularity Optimization algorithm groups the nodes in the graph by optimizing the graphs modularity.",
"signature": "gds.beta.modularityOptimization.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodes :: INTEGER?, didConverge :: BOOLEAN?, ranIterations :: INTEGER?, modularity :: FLOAT?, communityCount :: INTEGER?, communityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.modularityOptimization.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.modularityOptimization.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.modularityOptimization.stream",
"description": "The Modularity Optimization algorithm groups the nodes in the graph by optimizing the graphs modularity.",
"signature": "gds.beta.modularityOptimization.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, communityId :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.beta.modularityOptimization.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.modularityOptimization.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.modularityOptimization.write",
"description": "The Modularity Optimization algorithm groups the nodes in the graph by optimizing the graphs modularity.",
"signature": "gds.beta.modularityOptimization.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodes :: INTEGER?, didConverge :: BOOLEAN?, ranIterations :: INTEGER?, modularity :: FLOAT?, communityCount :: INTEGER?, communityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.modularityOptimization.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.modularityOptimization.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.node2vec.mutate",
"description": "The Node2Vec algorithm computes embeddings for nodes based on random walks.",
"signature": "gds.beta.node2vec.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeCount :: INTEGER?, nodePropertiesWritten :: INTEGER?, lossPerIteration :: LIST? OF FLOAT?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.node2vec.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.node2vec.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.node2vec.stream",
"description": "The Node2Vec algorithm computes embeddings for nodes based on random walks.",
"signature": "gds.beta.node2vec.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, embedding :: LIST? OF FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.node2vec.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.node2vec.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.node2vec.write",
"description": "The Node2Vec algorithm computes embeddings for nodes based on random walks.",
"signature": "gds.beta.node2vec.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeCount :: INTEGER?, nodePropertiesWritten :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, configuration :: MAP?, lossPerIteration :: LIST? OF FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.node2vec.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.beta.node2vec.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.drop",
"description": "Drops a pipeline and frees up the resources it occupies.",
"signature": "gds.beta.pipeline.drop(pipelineName :: STRING?, failIfMissing = true :: BOOLEAN?) :: (pipelineInfo :: MAP?, pipelineName :: STRING?, pipelineType :: STRING?, creationTime :: DATETIME?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.exists",
"description": "Checks if a given pipeline exists in the pipeline catalog.",
"signature": "gds.beta.pipeline.exists(pipelineName :: STRING?) :: (pipelineName :: STRING?, pipelineType :: STRING?, exists :: BOOLEAN?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.addFeature",
"description": "Add a feature step to an existing link prediction pipeline.",
"signature": "gds.beta.pipeline.linkPrediction.addFeature(pipelineName :: STRING?, featureType :: STRING?, configuration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureSteps :: LIST? OF MAP?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.addLogisticRegression",
"description": "Add a logistic regression configuration to the parameter space of the link prediction train pipeline.",
"signature": "gds.beta.pipeline.linkPrediction.addLogisticRegression(pipelineName :: STRING?, config = {} :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureSteps :: LIST? OF MAP?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.addNodeProperty",
"description": "Add a node property step to an existing link prediction pipeline.",
"signature": "gds.beta.pipeline.linkPrediction.addNodeProperty(pipelineName :: STRING?, procedureName :: STRING?, procedureConfiguration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureSteps :: LIST? OF MAP?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.configureSplit",
"description": "Configures the split of the link prediction pipeline.",
"signature": "gds.beta.pipeline.linkPrediction.configureSplit(pipelineName :: STRING?, configuration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureSteps :: LIST? OF MAP?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.create",
"description": "Creates a link prediction pipeline in the pipeline catalog.",
"signature": "gds.beta.pipeline.linkPrediction.create(pipelineName :: STRING?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureSteps :: LIST? OF MAP?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.predict.mutate",
"description": "Predicts relationships for all non-connected node pairs based on a previously trained LinkPrediction model.",
"signature": "gds.beta.pipeline.linkPrediction.predict.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, probabilityDistribution :: MAP?, samplingStats :: MAP?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.predict.mutate.estimate",
"description": "Estimates memory for predicting relationships for all non-connected node pairs based on a previously trained LinkPrediction model",
"signature": "gds.beta.pipeline.linkPrediction.predict.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.predict.stream",
"description": "Predicts relationships for all non-connected node pairs based on a previously trained LinkPrediction model.",
"signature": "gds.beta.pipeline.linkPrediction.predict.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (node1 :: INTEGER?, node2 :: INTEGER?, probability :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.predict.stream.estimate",
"description": "Estimates memory for predicting relationships for all non-connected node pairs based on a previously trained LinkPrediction model",
"signature": "gds.beta.pipeline.linkPrediction.predict.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.train",
"description": "Trains a link prediction model based on a pipeline",
"signature": "gds.beta.pipeline.linkPrediction.train(graphName :: STRING?, configuration = {} :: MAP?) :: (modelSelectionStats :: MAP?, trainMillis :: INTEGER?, modelInfo :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.linkPrediction.train.estimate",
"description": "Estimates memory for applying a linkPrediction model",
"signature": "gds.beta.pipeline.linkPrediction.train.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.list",
"description": "Lists all pipelines contained in the pipeline catalog.",
"signature": "gds.beta.pipeline.list(pipelineName = __NO_VALUE :: STRING?) :: (pipelineInfo :: MAP?, pipelineName :: STRING?, pipelineType :: STRING?, creationTime :: DATETIME?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.addLogisticRegression",
"description": "Add a logistic regression configuration to the parameter space of the node classification train pipeline.",
"signature": "gds.beta.pipeline.nodeClassification.addLogisticRegression(pipelineName :: STRING?, config = {} :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.addNodeProperty",
"description": "Add a node property step to an existing node classification training pipeline.",
"signature": "gds.beta.pipeline.nodeClassification.addNodeProperty(pipelineName :: STRING?, procedureName :: STRING?, procedureConfiguration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.configureSplit",
"description": "Configures the split of the node classification training pipeline.",
"signature": "gds.beta.pipeline.nodeClassification.configureSplit(pipelineName :: STRING?, configuration :: MAP?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.create",
"description": "Creates a node classification training pipeline in the pipeline catalog.",
"signature": "gds.beta.pipeline.nodeClassification.create(pipelineName :: STRING?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.predict.mutate",
"description": "Predicts classes for all nodes based on a previously trained pipeline model",
"signature": "gds.beta.pipeline.nodeClassification.predict.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.predict.mutate.estimate",
"description": "Estimates memory for predicting classes for all nodes based on a previously trained pipeline model",
"signature": "gds.beta.pipeline.nodeClassification.predict.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.predict.stream",
"description": "Predicts classes for all nodes based on a previously trained pipeline model",
"signature": "gds.beta.pipeline.nodeClassification.predict.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, predictedClass :: INTEGER?, predictedProbabilities :: LIST? OF FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.predict.stream.estimate",
"description": "Estimates memory for predicting classes for all nodes based on a previously trained pipeline model",
"signature": "gds.beta.pipeline.nodeClassification.predict.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.predict.write",
"description": "Predicts classes for all nodes based on a previously trained pipeline model",
"signature": "gds.beta.pipeline.nodeClassification.predict.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.predict.write.estimate",
"description": "Estimates memory for predicting classes for all nodes based on a previously trained pipeline model",
"signature": "gds.beta.pipeline.nodeClassification.predict.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.selectFeatures",
"description": "Add one or several features to an existing node classification training pipeline.",
"signature": "gds.beta.pipeline.nodeClassification.selectFeatures(pipelineName :: STRING?, nodeProperties :: ANY?) :: (name :: STRING?, nodePropertySteps :: LIST? OF MAP?, featureProperties :: LIST? OF STRING?, splitConfig :: MAP?, autoTuningConfig :: MAP?, parameterSpace :: ANY?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.train",
"description": "Trains a node classification model based on a pipeline",
"signature": "gds.beta.pipeline.nodeClassification.train(graphName :: STRING?, configuration = {} :: MAP?) :: (modelSelectionStats :: MAP?, trainMillis :: INTEGER?, modelInfo :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.beta.pipeline.nodeClassification.train.estimate",
"description": "Estimates memory for training a node classification model based on a pipeline",
"signature": "gds.beta.pipeline.nodeClassification.train.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.betweenness.mutate",
"description": "Betweenness centrality measures the relative information flow that passes through a node.",
"signature": "gds.betweenness.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, mutateMillis :: INTEGER?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.betweenness.mutate.estimate",
"description": "Betweenness centrality measures the relative information flow that passes through a node.",
"signature": "gds.betweenness.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.betweenness.stats",
"description": "Betweenness centrality measures the relative information flow that passes through a node.",
"signature": "gds.betweenness.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.betweenness.stats.estimate",
"description": "Betweenness centrality measures the relative information flow that passes through a node.",
"signature": "gds.betweenness.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.betweenness.stream",
"description": "Betweenness centrality measures the relative information flow that passes through a node.",
"signature": "gds.betweenness.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, score :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.betweenness.stream.estimate",
"description": "Betweenness centrality measures the relative information flow that passes through a node.",
"signature": "gds.betweenness.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.betweenness.write",
"description": "Betweenness centrality measures the relative information flow that passes through a node.",
"signature": "gds.betweenness.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, writeMillis :: INTEGER?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.betweenness.write.estimate",
"description": "Betweenness centrality measures the relative information flow that passes through a node.",
"signature": "gds.betweenness.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.bfs.mutate",
"description": "BFS is a traversal algorithm, which explores all of the neighbor nodes at the present depth prior to moving on to the nodes at the next depth level.",
"signature": "gds.bfs.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.bfs.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.bfs.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.bfs.stats",
"description": "BFS is a traversal algorithm, which explores all of the neighbor nodes at the present depth prior to moving on to the nodes at the next depth level.",
"signature": "gds.bfs.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.bfs.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.bfs.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.bfs.stream",
"description": "BFS is a traversal algorithm, which explores all of the neighbor nodes at the present depth prior to moving on to the nodes at the next depth level.",
"signature": "gds.bfs.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (sourceNode :: INTEGER?, nodeIds :: LIST? OF INTEGER?, path :: PATH?)",
"type": "procedure"
},
{
"name": "gds.bfs.stream.estimate",
"description": "BFS is a traversal algorithm, which explores all of the neighbor nodes at the present depth prior to moving on to the nodes at the next depth level.",
"signature": "gds.bfs.stream.estimate(graphName :: ANY?, configuration = {} :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.debug.sysInfo",
"description": "Returns details about the status of the system",
"signature": "gds.debug.sysInfo() :: (key :: STRING?, value :: ANY?)",
"type": "procedure"
},
{
"name": "gds.degree.mutate",
"description": "Degree centrality measures the number of incoming and outgoing relationships from a node.",
"signature": "gds.degree.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, centralityDistribution :: MAP?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.degree.mutate.estimate",
"description": "Degree centrality measures the number of incoming and outgoing relationships from a node.",
"signature": "gds.degree.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.degree.stats",
"description": "Degree centrality measures the number of incoming and outgoing relationships from a node.",
"signature": "gds.degree.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.degree.stats.estimate",
"description": "Degree centrality measures the number of incoming and outgoing relationships from a node.",
"signature": "gds.degree.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.degree.stream",
"description": "Degree centrality measures the number of incoming and outgoing relationships from a node.",
"signature": "gds.degree.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, score :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.degree.stream.estimate",
"description": "Degree centrality measures the number of incoming and outgoing relationships from a node.",
"signature": "gds.degree.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.degree.write",
"description": "Degree centrality measures the number of incoming and outgoing relationships from a node.",
"signature": "gds.degree.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, centralityDistribution :: MAP?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.degree.write.estimate",
"description": "Degree centrality measures the number of incoming and outgoing relationships from a node.",
"signature": "gds.degree.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.dfs.mutate",
"description": "Depth-first search (DFS) is an algorithm for traversing or searching tree or graph data structures. The algorithm starts at the root node (selecting some arbitrary node as the root node in the case of a graph) and explores as far as possible along each branch before backtracking.",
"signature": "gds.dfs.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.dfs.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.dfs.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.dfs.stream",
"description": "Depth-first search (DFS) is an algorithm for traversing or searching tree or graph data structures. The algorithm starts at the root node (selecting some arbitrary node as the root node in the case of a graph) and explores as far as possible along each branch before backtracking.",
"signature": "gds.dfs.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (sourceNode :: INTEGER?, nodeIds :: LIST? OF INTEGER?, path :: PATH?)",
"type": "procedure"
},
{
"name": "gds.dfs.stream.estimate",
"description": "Depth-first search (DFS) is an algorithm for traversing or searching tree or graph data structures. The algorithm starts at the root node (selecting some arbitrary node as the root node in the case of a graph) and explores as far as possible along each branch before backtracking.",
"signature": "gds.dfs.stream.estimate(graphName :: ANY?, configuration = {} :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.eigenvector.mutate",
"description": "Eigenvector Centrality is an algorithm that measures the transitive influence or connectivity of nodes.",
"signature": "gds.eigenvector.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.eigenvector.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.eigenvector.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.eigenvector.stats",
"description": "Eigenvector Centrality is an algorithm that measures the transitive influence or connectivity of nodes.",
"signature": "gds.eigenvector.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.eigenvector.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.eigenvector.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.eigenvector.stream",
"description": "Eigenvector Centrality is an algorithm that measures the transitive influence or connectivity of nodes.",
"signature": "gds.eigenvector.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, score :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.eigenvector.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.eigenvector.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.eigenvector.write",
"description": "Eigenvector Centrality is an algorithm that measures the transitive influence or connectivity of nodes.",
"signature": "gds.eigenvector.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.eigenvector.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.eigenvector.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.fastRP.mutate",
"description": "Random Projection produces node embeddings via the fastrp algorithm",
"signature": "gds.fastRP.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (nodePropertiesWritten :: INTEGER?, mutateMillis :: INTEGER?, nodeCount :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.fastRP.mutate.estimate",
"description": "Random Projection produces node embeddings via the fastrp algorithm",
"signature": "gds.fastRP.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.fastRP.stats",
"description": "Random Projection produces node embeddings via the fastrp algorithm",
"signature": "gds.fastRP.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeCount :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.fastRP.stats.estimate",
"description": "Random Projection produces node embeddings via the fastrp algorithm",
"signature": "gds.fastRP.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.fastRP.stream",
"description": "Random Projection produces node embeddings via the fastrp algorithm",
"signature": "gds.fastRP.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, embedding :: LIST? OF FLOAT?)",
"type": "procedure"
},
{
"name": "gds.fastRP.stream.estimate",
"description": "Random Projection produces node embeddings via the fastrp algorithm",
"signature": "gds.fastRP.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.fastRP.write",
"description": "Random Projection produces node embeddings via the fastrp algorithm",
"signature": "gds.fastRP.write(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeCount :: INTEGER?, nodePropertiesWritten :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.fastRP.write.estimate",
"description": "Random Projection produces node embeddings via the fastrp algorithm",
"signature": "gds.fastRP.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.graph.deleteRelationships",
"description": "Delete the relationship type for a given graph stored in the graph-catalog.",
"signature": "gds.graph.deleteRelationships(graphName :: STRING?, relationshipType :: STRING?) :: (graphName :: STRING?, relationshipType :: STRING?, deletedRelationships :: INTEGER?, deletedProperties :: MAP?)",
"type": "procedure"
},
{
"name": "gds.graph.drop",
"description": "Drops a named graph from the catalog and frees up the resources it occupies.",
"signature": "gds.graph.drop(graphName :: ANY?, failIfMissing = true :: BOOLEAN?, dbName = :: STRING?, username = :: STRING?) :: (graphName :: STRING?, database :: STRING?, memoryUsage :: STRING?, sizeInBytes :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, configuration :: MAP?, density :: FLOAT?, creationTime :: DATETIME?, modificationTime :: DATETIME?, schema :: MAP?)",
"type": "procedure"
},
{
"name": "gds.graph.exists",
"description": "Checks if a graph exists in the catalog.",
"signature": "gds.graph.exists(graphName :: STRING?) :: (graphName :: STRING?, exists :: BOOLEAN?)",
"type": "procedure"
},
{
"name": "gds.graph.export",
"description": "Exports a named graph into a new offline Neo4j database.",
"signature": "gds.graph.export(graphName :: STRING?, configuration = {} :: MAP?) :: (dbName :: STRING?, graphName :: STRING?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, relationshipTypeCount :: INTEGER?, nodePropertyCount :: INTEGER?, relationshipPropertyCount :: INTEGER?, writeMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.graph.list",
"description": "Lists information about named graphs stored in the catalog.",
"signature": "gds.graph.list(graphName = __NO_VALUE :: STRING?) :: (degreeDistribution :: MAP?, graphName :: STRING?, database :: STRING?, memoryUsage :: STRING?, sizeInBytes :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, configuration :: MAP?, density :: FLOAT?, creationTime :: DATETIME?, modificationTime :: DATETIME?, schema :: MAP?)",
"type": "procedure"
},
{
"name": "gds.graph.nodeProperties.drop",
"description": "Removes node properties from a projected graph.",
"signature": "gds.graph.nodeProperties.drop(graphName :: STRING?, nodeProperties :: ANY?, configuration = {} :: MAP?) :: (graphName :: STRING?, nodeProperties :: LIST? OF STRING?, propertiesRemoved :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.graph.nodeProperties.stream",
"description": "Streams the given node properties.",
"signature": "gds.graph.nodeProperties.stream(graphName :: STRING?, nodeProperties :: ANY?, nodeLabels = [*] :: ANY?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, nodeProperty :: STRING?, propertyValue :: ANY?)",
"type": "procedure"
},
{
"name": "gds.graph.nodeProperties.write",
"description": "Writes the given node properties to an online Neo4j database.",
"signature": "gds.graph.nodeProperties.write(graphName :: STRING?, nodeProperties :: ANY?, nodeLabels = [*] :: ANY?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, graphName :: STRING?, nodeProperties :: LIST? OF STRING?, propertiesWritten :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.graph.nodeProperty.stream",
"description": "Streams the given node property.",
"signature": "gds.graph.nodeProperty.stream(graphName :: STRING?, nodeProperties :: STRING?, nodeLabels = [*] :: ANY?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, propertyValue :: ANY?)",
"type": "procedure"
},
{
"name": "gds.graph.project",
"description": "Creates a named graph in the catalog for use by algorithms.",
"signature": "gds.graph.project(graphName :: STRING?, nodeProjection :: ANY?, relationshipProjection :: ANY?, configuration = {} :: MAP?) :: (nodeProjection :: MAP?, relationshipProjection :: MAP?, graphName :: STRING?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, projectMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.graph.project.cypher",
"description": "Creates a named graph in the catalog for use by algorithms.",
"signature": "gds.graph.project.cypher(graphName :: STRING?, nodeQuery :: STRING?, relationshipQuery :: STRING?, configuration = {} :: MAP?) :: (nodeQuery :: STRING?, relationshipQuery :: STRING?, graphName :: STRING?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, projectMillis :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.graph.project.cypher.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.graph.project.cypher.estimate(nodeQuery :: STRING?, relationshipQuery :: STRING?, configuration = {} :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.graph.project.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.graph.project.estimate(nodeProjection :: ANY?, relationshipProjection :: ANY?, configuration = {} :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.graph.relationship.write",
"description": "Writes the given relationship and an optional relationship property to an online Neo4j database.",
"signature": "gds.graph.relationship.write(graphName :: STRING?, relationshipType :: STRING?, relationshipProperty = :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, graphName :: STRING?, relationshipType :: STRING?, relationshipProperty :: STRING?, relationshipsWritten :: INTEGER?, propertiesWritten :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.graph.relationshipProperties.stream",
"description": "Streams the given relationship properties.",
"signature": "gds.graph.relationshipProperties.stream(graphName :: STRING?, relationshipProperties :: LIST? OF STRING?, relationshipTypes = [*] :: LIST? OF STRING?, configuration = {} :: MAP?) :: (sourceNodeId :: INTEGER?, targetNodeId :: INTEGER?, relationshipType :: STRING?, relationshipProperty :: STRING?, propertyValue :: NUMBER?)",
"type": "procedure"
},
{
"name": "gds.graph.relationshipProperty.stream",
"description": "Streams the given relationship property.",
"signature": "gds.graph.relationshipProperty.stream(graphName :: STRING?, relationshipProperty :: STRING?, relationshipTypes = [*] :: LIST? OF STRING?, configuration = {} :: MAP?) :: (sourceNodeId :: INTEGER?, targetNodeId :: INTEGER?, relationshipType :: STRING?, propertyValue :: NUMBER?)",
"type": "procedure"
},
{
"name": "gds.graph.relationships.drop",
"description": "Delete the relationship type for a given graph stored in the graph-catalog.",
"signature": "gds.graph.relationships.drop(graphName :: STRING?, relationshipType :: STRING?) :: (graphName :: STRING?, relationshipType :: STRING?, deletedRelationships :: INTEGER?, deletedProperties :: MAP?)",
"type": "procedure"
},
{
"name": "gds.graph.removeNodeProperties",
"description": "Removes node properties from a projected graph.",
"signature": "gds.graph.removeNodeProperties(graphName :: STRING?, nodeProperties :: ANY?, configuration = {} :: MAP?) :: (graphName :: STRING?, nodeProperties :: LIST? OF STRING?, propertiesRemoved :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.graph.streamNodeProperties",
"description": "Streams the given node properties.",
"signature": "gds.graph.streamNodeProperties(graphName :: STRING?, nodeProperties :: ANY?, nodeLabels = [*] :: ANY?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, nodeProperty :: STRING?, propertyValue :: ANY?)",
"type": "procedure"
},
{
"name": "gds.graph.streamNodeProperty",
"description": "Streams the given node property.",
"signature": "gds.graph.streamNodeProperty(graphName :: STRING?, nodeProperties :: STRING?, nodeLabels = [*] :: ANY?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, propertyValue :: ANY?)",
"type": "procedure"
},
{
"name": "gds.graph.streamRelationshipProperties",
"description": "Streams the given relationship properties.",
"signature": "gds.graph.streamRelationshipProperties(graphName :: STRING?, relationshipProperties :: LIST? OF STRING?, relationshipTypes = [*] :: LIST? OF STRING?, configuration = {} :: MAP?) :: (sourceNodeId :: INTEGER?, targetNodeId :: INTEGER?, relationshipType :: STRING?, relationshipProperty :: STRING?, propertyValue :: NUMBER?)",
"type": "procedure"
},
{
"name": "gds.graph.streamRelationshipProperty",
"description": "Streams the given relationship property.",
"signature": "gds.graph.streamRelationshipProperty(graphName :: STRING?, relationshipProperties :: STRING?, relationshipTypes = [*] :: LIST? OF STRING?, configuration = {} :: MAP?) :: (sourceNodeId :: INTEGER?, targetNodeId :: INTEGER?, relationshipType :: STRING?, propertyValue :: NUMBER?)",
"type": "procedure"
},
{
"name": "gds.graph.writeNodeProperties",
"description": "Writes the given node properties to an online Neo4j database.",
"signature": "gds.graph.writeNodeProperties(graphName :: STRING?, nodeProperties :: ANY?, nodeLabels = [*] :: ANY?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, graphName :: STRING?, nodeProperties :: LIST? OF STRING?, propertiesWritten :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.graph.writeRelationship",
"description": "Writes the given relationship and an optional relationship property to an online Neo4j database.",
"signature": "gds.graph.writeRelationship(graphName :: STRING?, relationshipType :: STRING?, relationshipProperty = :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, graphName :: STRING?, relationshipType :: STRING?, relationshipProperty :: STRING?, relationshipsWritten :: INTEGER?, propertiesWritten :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.knn.mutate",
"description": "The k-nearest neighbor graph algorithm constructs relationships between nodes if the distance between two nodes is among the k nearest distances compared to other nodes.KNN computes distances based on the similarity of node properties",
"signature": "gds.knn.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, nodePairsConsidered :: INTEGER?, didConverge :: BOOLEAN?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, relationshipsWritten :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.knn.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.knn.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.knn.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.knn.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, nodePairsConsidered :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, similarityPairs :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.knn.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.knn.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.knn.stream",
"description": "The k-nearest neighbor graph algorithm constructs relationships between nodes if the distance between two nodes is among the k nearest distances compared to other nodes.KNN computes distances based on the similarity of node properties",
"signature": "gds.knn.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (node1 :: INTEGER?, node2 :: INTEGER?, similarity :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.knn.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.knn.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.knn.write",
"description": "The k-nearest neighbor graph algorithm constructs relationships between nodes if the distance between two nodes is among the k nearest distances compared to other nodes.KNN computes distances based on the similarity of node properties",
"signature": "gds.knn.write(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, nodePairsConsidered :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, relationshipsWritten :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.knn.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.knn.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.labelPropagation.mutate",
"description": "The Label Propagation algorithm is a fast algorithm for finding communities in a graph.",
"signature": "gds.labelPropagation.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, communityCount :: INTEGER?, communityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.labelPropagation.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.labelPropagation.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.labelPropagation.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.labelPropagation.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, communityCount :: INTEGER?, communityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.labelPropagation.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.labelPropagation.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.labelPropagation.stream",
"description": "The Label Propagation algorithm is a fast algorithm for finding communities in a graph.",
"signature": "gds.labelPropagation.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, communityId :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.labelPropagation.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.labelPropagation.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.labelPropagation.write",
"description": "The Label Propagation algorithm is a fast algorithm for finding communities in a graph.",
"signature": "gds.labelPropagation.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, communityCount :: INTEGER?, communityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.labelPropagation.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.labelPropagation.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.localClusteringCoefficient.mutate",
"description": "The local clustering coefficient is a metric quantifying how connected the neighborhood of a node is.",
"signature": "gds.localClusteringCoefficient.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, averageClusteringCoefficient :: FLOAT?, nodeCount :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.localClusteringCoefficient.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.localClusteringCoefficient.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.localClusteringCoefficient.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.localClusteringCoefficient.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (averageClusteringCoefficient :: FLOAT?, nodeCount :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.localClusteringCoefficient.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.localClusteringCoefficient.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.localClusteringCoefficient.stream",
"description": "The local clustering coefficient is a metric quantifying how connected the neighborhood of a node is.",
"signature": "gds.localClusteringCoefficient.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, localClusteringCoefficient :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.localClusteringCoefficient.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.localClusteringCoefficient.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.localClusteringCoefficient.write",
"description": "The local clustering coefficient is a metric quantifying how connected the neighborhood of a node is.",
"signature": "gds.localClusteringCoefficient.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, averageClusteringCoefficient :: FLOAT?, nodeCount :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.localClusteringCoefficient.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.localClusteringCoefficient.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.louvain.mutate",
"description": "The Louvain method for community detection is an algorithm for detecting communities in networks.",
"signature": "gds.louvain.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, modularity :: FLOAT?, modularities :: LIST? OF FLOAT?, ranLevels :: INTEGER?, communityCount :: INTEGER?, communityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.louvain.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.louvain.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.louvain.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.louvain.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (modularity :: FLOAT?, modularities :: LIST? OF FLOAT?, ranLevels :: INTEGER?, communityCount :: INTEGER?, communityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.louvain.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.louvain.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.louvain.stream",
"description": "The Louvain method for community detection is an algorithm for detecting communities in networks.",
"signature": "gds.louvain.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, communityId :: INTEGER?, intermediateCommunityIds :: LIST? OF INTEGER?)",
"type": "procedure"
},
{
"name": "gds.louvain.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.louvain.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.louvain.write",
"description": "The Louvain method for community detection is an algorithm for detecting communities in networks.",
"signature": "gds.louvain.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, modularity :: FLOAT?, modularities :: LIST? OF FLOAT?, ranLevels :: INTEGER?, communityCount :: INTEGER?, communityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.louvain.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.louvain.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.nodeSimilarity.mutate",
"description": "The Node Similarity algorithm compares a set of nodes based on the nodes they are connected to. Two nodes are considered similar if they share many of the same neighbors. Node Similarity computes pair-wise similarities based on the Jaccard metric.",
"signature": "gds.nodeSimilarity.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, relationshipsWritten :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.nodeSimilarity.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.nodeSimilarity.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.nodeSimilarity.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.nodeSimilarity.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, similarityPairs :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.nodeSimilarity.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.nodeSimilarity.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.nodeSimilarity.stream",
"description": "The Node Similarity algorithm compares a set of nodes based on the nodes they are connected to. Two nodes are considered similar if they share many of the same neighbors. Node Similarity computes pair-wise similarities based on the Jaccard metric.",
"signature": "gds.nodeSimilarity.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (node1 :: INTEGER?, node2 :: INTEGER?, similarity :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.nodeSimilarity.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.nodeSimilarity.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.nodeSimilarity.write",
"description": "The Node Similarity algorithm compares a set of nodes based on the nodes they are connected to. Two nodes are considered similar if they share many of the same neighbors. Node Similarity computes pair-wise similarities based on the Jaccard metric.",
"signature": "gds.nodeSimilarity.write(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, nodesCompared :: INTEGER?, relationshipsWritten :: INTEGER?, similarityDistribution :: MAP?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.nodeSimilarity.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.nodeSimilarity.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.pageRank.mutate",
"description": "Page Rank is an algorithm that measures the transitive influence or connectivity of nodes.",
"signature": "gds.pageRank.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.pageRank.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.pageRank.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.pageRank.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.pageRank.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.pageRank.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.pageRank.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.pageRank.stream",
"description": "Page Rank is an algorithm that measures the transitive influence or connectivity of nodes.",
"signature": "gds.pageRank.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, score :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.pageRank.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.pageRank.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.pageRank.write",
"description": "Page Rank is an algorithm that measures the transitive influence or connectivity of nodes.",
"signature": "gds.pageRank.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, ranIterations :: INTEGER?, didConverge :: BOOLEAN?, centralityDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.pageRank.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.pageRank.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.randomWalk.stats",
"description": "Random Walk is an algorithm that provides random paths in a graph. It’s similar to how a drunk person traverses a city.",
"signature": "gds.randomWalk.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.randomWalk.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.randomWalk.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.randomWalk.stream",
"description": "Random Walk is an algorithm that provides random paths in a graph. It’s similar to how a drunk person traverses a city.",
"signature": "gds.randomWalk.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeIds :: LIST? OF INTEGER?, path :: PATH?)",
"type": "procedure"
},
{
"name": "gds.randomWalk.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.randomWalk.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.astar.mutate",
"description": "The A* shortest path algorithm computes the shortest path between a pair of nodes. It uses the relationship weight property to compare path lengths. In addition, this implementation uses the haversine distance as a heuristic to converge faster.",
"signature": "gds.shortestPath.astar.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.astar.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.astar.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.astar.stream",
"description": "The A* shortest path algorithm computes the shortest path between a pair of nodes. It uses the relationship weight property to compare path lengths. In addition, this implementation uses the haversine distance as a heuristic to converge faster.",
"signature": "gds.shortestPath.astar.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (index :: INTEGER?, sourceNode :: INTEGER?, targetNode :: INTEGER?, totalCost :: FLOAT?, nodeIds :: LIST? OF INTEGER?, costs :: LIST? OF FLOAT?, path :: PATH?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.astar.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.astar.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.astar.write",
"description": "The A* shortest path algorithm computes the shortest path between a pair of nodes. It uses the relationship weight property to compare path lengths. In addition, this implementation uses the haversine distance as a heuristic to converge faster.",
"signature": "gds.shortestPath.astar.write(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.astar.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.astar.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.dijkstra.mutate",
"description": "The Dijkstra shortest path algorithm computes the shortest (weighted) path between a pair of nodes.",
"signature": "gds.shortestPath.dijkstra.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.dijkstra.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.dijkstra.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.dijkstra.stream",
"description": "The Dijkstra shortest path algorithm computes the shortest (weighted) path between a pair of nodes.",
"signature": "gds.shortestPath.dijkstra.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (index :: INTEGER?, sourceNode :: INTEGER?, targetNode :: INTEGER?, totalCost :: FLOAT?, nodeIds :: LIST? OF INTEGER?, costs :: LIST? OF FLOAT?, path :: PATH?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.dijkstra.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.dijkstra.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.dijkstra.write",
"description": "The Dijkstra shortest path algorithm computes the shortest (weighted) path between a pair of nodes.",
"signature": "gds.shortestPath.dijkstra.write(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.dijkstra.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.dijkstra.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.yens.mutate",
"description": "The Yen's shortest path algorithm computes the k shortest (weighted) paths between a pair of nodes.",
"signature": "gds.shortestPath.yens.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, mutateMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.yens.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.yens.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.yens.stream",
"description": "The Yen's shortest path algorithm computes the k shortest (weighted) paths between a pair of nodes.",
"signature": "gds.shortestPath.yens.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (index :: INTEGER?, sourceNode :: INTEGER?, targetNode :: INTEGER?, totalCost :: FLOAT?, nodeIds :: LIST? OF INTEGER?, costs :: LIST? OF FLOAT?, path :: PATH?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.yens.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.yens.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.yens.write",
"description": "The Yen's shortest path algorithm computes the k shortest (weighted) paths between a pair of nodes.",
"signature": "gds.shortestPath.yens.write(graphName :: STRING?, configuration = {} :: MAP?) :: (relationshipsWritten :: INTEGER?, writeMillis :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.shortestPath.yens.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.shortestPath.yens.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.triangleCount.mutate",
"description": "Triangle counting is a community detection graph algorithm that is used to determine the number of triangles passing through each node in the graph.",
"signature": "gds.triangleCount.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, globalTriangleCount :: INTEGER?, nodeCount :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.triangleCount.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.triangleCount.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.triangleCount.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.triangleCount.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (globalTriangleCount :: INTEGER?, nodeCount :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.triangleCount.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.triangleCount.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.triangleCount.stream",
"description": "Triangle counting is a community detection graph algorithm that is used to determine the number of triangles passing through each node in the graph.",
"signature": "gds.triangleCount.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, triangleCount :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.triangleCount.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.triangleCount.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.triangleCount.write",
"description": "Triangle counting is a community detection graph algorithm that is used to determine the number of triangles passing through each node in the graph.",
"signature": "gds.triangleCount.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, globalTriangleCount :: INTEGER?, nodeCount :: INTEGER?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.triangleCount.write.estimate",
"description": "Triangle counting is a community detection graph algorithm that is used to determine the number of triangles passing through each node in the graph.",
"signature": "gds.triangleCount.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.wcc.mutate",
"description": "The WCC algorithm finds sets of connected nodes in an undirected graph, where all nodes in the same set form a connected component.",
"signature": "gds.wcc.mutate(graphName :: STRING?, configuration = {} :: MAP?) :: (mutateMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, componentCount :: INTEGER?, componentDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.wcc.mutate.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.wcc.mutate.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.wcc.stats",
"description": "Executes the algorithm and returns result statistics without writing the result to Neo4j.",
"signature": "gds.wcc.stats(graphName :: STRING?, configuration = {} :: MAP?) :: (componentCount :: INTEGER?, componentDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.wcc.stats.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.wcc.stats.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.wcc.stream",
"description": "The WCC algorithm finds sets of connected nodes in an undirected graph, where all nodes in the same set form a connected component.",
"signature": "gds.wcc.stream(graphName :: STRING?, configuration = {} :: MAP?) :: (nodeId :: INTEGER?, componentId :: INTEGER?)",
"type": "procedure"
},
{
"name": "gds.wcc.stream.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.wcc.stream.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.wcc.write",
"description": "The WCC algorithm finds sets of connected nodes in an undirected graph, where all nodes in the same set form a connected component.",
"signature": "gds.wcc.write(graphName :: STRING?, configuration = {} :: MAP?) :: (writeMillis :: INTEGER?, nodePropertiesWritten :: INTEGER?, componentCount :: INTEGER?, componentDistribution :: MAP?, postProcessingMillis :: INTEGER?, preProcessingMillis :: INTEGER?, computeMillis :: INTEGER?, configuration :: MAP?)",
"type": "procedure"
},
{
"name": "gds.wcc.write.estimate",
"description": "Returns an estimation of the memory consumption for that procedure.",
"signature": "gds.wcc.write.estimate(graphNameOrConfiguration :: ANY?, algoConfiguration :: MAP?) :: (requiredMemory :: STRING?, treeView :: STRING?, mapView :: MAP?, bytesMin :: INTEGER?, bytesMax :: INTEGER?, nodeCount :: INTEGER?, relationshipCount :: INTEGER?, heapPercentageMin :: FLOAT?, heapPercentageMax :: FLOAT?)",
"type": "procedure"
},
{
"name": "gds.alpha.graph.project",
"description": "Creates a named graph in the catalog for use by algorithms.",
"signature": "gds.alpha.graph.project(graphName :: STRING?, sourceNode :: ANY?, targetNode = null :: ANY?, nodesConfig = null :: MAP?, relationshipConfig = null :: MAP?) :: (MAP?)",
"type": "function"
},
{
"name": "gds.alpha.linkprediction.adamicAdar",
"description": "Given two nodes, calculate Adamic Adar similarity",
"signature": "gds.alpha.linkprediction.adamicAdar(node1 :: NODE?, node2 :: NODE?, config = {} :: MAP?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.alpha.linkprediction.commonNeighbors",
"description": "Given two nodes, returns the number of common neighbors",
"signature": "gds.alpha.linkprediction.commonNeighbors(node1 :: NODE?, node2 :: NODE?, config = {} :: MAP?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.alpha.linkprediction.preferentialAttachment",
"description": "Given two nodes, calculate Preferential Attachment",
"signature": "gds.alpha.linkprediction.preferentialAttachment(node1 :: NODE?, node2 :: NODE?, config = {} :: MAP?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.alpha.linkprediction.resourceAllocation",
"description": "Given two nodes, calculate Resource Allocation similarity",
"signature": "gds.alpha.linkprediction.resourceAllocation(node1 :: NODE?, node2 :: NODE?, config = {} :: MAP?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.alpha.linkprediction.sameCommunity",
"description": "Given two nodes, indicates if they have the same community",
"signature": "gds.alpha.linkprediction.sameCommunity(node1 :: NODE?, node2 :: NODE?, communityProperty = community :: STRING?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.alpha.linkprediction.totalNeighbors",
"description": "Given two nodes, calculate Total Neighbors",
"signature": "gds.alpha.linkprediction.totalNeighbors(node1 :: NODE?, node2 :: NODE?, config = {} :: MAP?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.alpha.ml.oneHotEncoding",
"description": "RETURN gds.alpha.ml.oneHotEncoding(availableValues, selectedValues) - return a list of selected values in a one hot encoding format.",
"signature": "gds.alpha.ml.oneHotEncoding(availableValues :: LIST? OF ANY?, selectedValues :: LIST? OF ANY?) :: (LIST? OF ANY?)",
"type": "function"
},
{
"name": "gds.graph.exists",
"description": "Checks if a graph exists in the catalog.",
"signature": "gds.graph.exists(graphName :: STRING?) :: (BOOLEAN?)",
"type": "function"
},
{
"name": "gds.similarity.cosine",
"description": "RETURN gds.similarity.cosine(vector1, vector2) - Given two collection vectors, calculate cosine similarity",
"signature": "gds.similarity.cosine(vector1 :: LIST? OF NUMBER?, vector2 :: LIST? OF NUMBER?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.similarity.euclidean",
"description": "RETURN gds.similarity.euclidean(vector1, vector2) - Given two collection vectors, calculate similarity based on euclidean distance",
"signature": "gds.similarity.euclidean(vector1 :: LIST? OF NUMBER?, vector2 :: LIST? OF NUMBER?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.similarity.euclideanDistance",
"description": "RETURN gds.similarity.euclideanDistance(vector1, vector2) - Given two collection vectors, calculate the euclidean distance (square root of the sum of the squared differences)",
"signature": "gds.similarity.euclideanDistance(vector1 :: LIST? OF NUMBER?, vector2 :: LIST? OF NUMBER?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.similarity.jaccard",
"description": "RETURN gds.similarity.jaccard(vector1, vector2) - Given two collection vectors, calculate Jaccard similarity",
"signature": "gds.similarity.jaccard(vector1 :: LIST? OF NUMBER?, vector2 :: LIST? OF NUMBER?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.similarity.overlap",
"description": "RETURN gds.similarity.overlap(vector1, vector2) - Given two collection vectors, calculate overlap similarity",
"signature": "gds.similarity.overlap(vector1 :: LIST? OF NUMBER?, vector2 :: LIST? OF NUMBER?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.similarity.pearson",
"description": "RETURN gds.similarity.pearson(vector1, vector2) - Given two collection vectors, calculate pearson similarity",
"signature": "gds.similarity.pearson(vector1 :: LIST? OF NUMBER?, vector2 :: LIST? OF NUMBER?) :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.util.NaN",
"description": "RETURN gds.util.NaN() - Returns NaN as a Cypher value.",
"signature": "gds.util.NaN() :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.util.asNode",
"description": "RETURN gds.util.asNode(nodeId) - Return the node objects for the given node id or null if none exists.",
"signature": "gds.util.asNode(nodeId :: NUMBER?) :: (NODE?)",
"type": "function"
},
{
"name": "gds.util.asNodes",
"description": "RETURN gds.util.asNodes(nodeIds) - Return the node objects for the given node ids or an empty list if none exists.",
"signature": "gds.util.asNodes(nodeIds :: LIST? OF NUMBER?) :: (LIST? OF ANY?)",
"type": "function"
},
{
"name": "gds.util.infinity",
"description": "RETURN gds.util.infinity() - Return infinity as a Cypher value.",
"signature": "gds.util.infinity() :: (FLOAT?)",
"type": "function"
},
{
"name": "gds.util.isFinite",
"description": "RETURN gds.util.isFinite(value) - Return true iff the given argument is a finite value (not ±Infinity, NaN, or null).",
"signature": "gds.util.isFinite(value :: NUMBER?) :: (BOOLEAN?)",
"type": "function"
},
{
"name": "gds.util.isInfinite",
"description": "RETURN gds.util.isInfinite(value) - Return true iff the given argument is not a finite value (not ±Infinity, NaN, or null).",
"signature": "gds.util.isInfinite(value :: NUMBER?) :: (BOOLEAN?)",
"type": "function"
},
{
"name": "gds.util.nodeProperty",
"description": "Returns a node property value from a named in-memory graph.",
"signature": "gds.util.nodeProperty(graphName :: STRING?, nodeId :: NUMBER?, propertyKey :: STRING?, nodeLabel = * :: STRING?) :: (ANY?)",
"type": "function"
},
{
"name": "gds.version",
"description": "RETURN gds.version() | Return the installed graph data science library version.",
"signature": "gds.version() :: (STRING?)",
"type": "function"
}
]